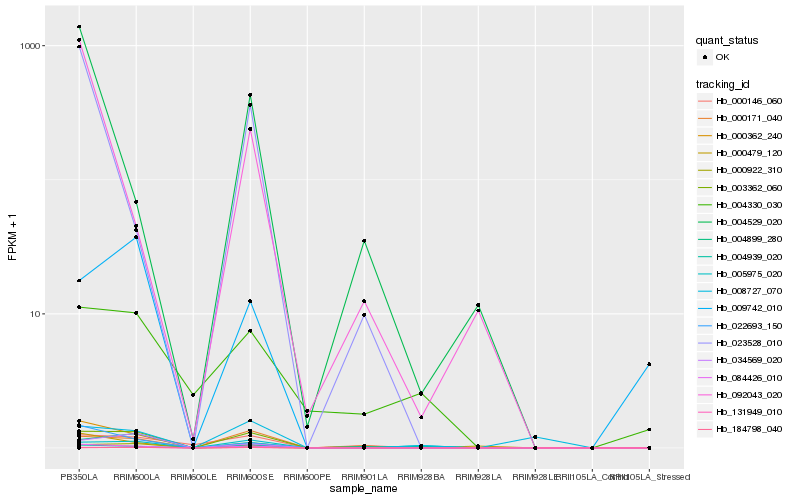
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_084426_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_003362_060 |
0.1331887831 |
- |
- |
- |
| 3 |
Hb_131949_010 |
0.1423587543 |
- |
- |
PREDICTED: uncharacterized protein LOC104227863, partial [Nicotiana sylvestris] |
| 4 |
Hb_022693_150 |
0.1529773892 |
- |
- |
- |
| 5 |
Hb_000171_040 |
0.2072111487 |
- |
- |
- |
| 6 |
Hb_000922_310 |
0.2663250137 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB98-like [Jatropha curcas] |
| 7 |
Hb_000362_240 |
0.2698966902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000146_060 |
0.2797527274 |
- |
- |
- |
| 9 |
Hb_034569_020 |
0.2816720024 |
- |
- |
- |
| 10 |
Hb_184798_040 |
0.2895999542 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 11 |
Hb_004899_280 |
0.2926803 |
- |
- |
- |
| 12 |
Hb_005975_020 |
0.2947695293 |
- |
- |
- |
| 13 |
Hb_023528_010 |
0.2953486132 |
- |
- |
hypothetical protein POPTR_0013s01580g [Populus trichocarpa] |
| 14 |
Hb_004529_020 |
0.3040879774 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 15 |
Hb_004330_030 |
0.3124588383 |
- |
- |
- |
| 16 |
Hb_000479_120 |
0.3159206799 |
transcription factor |
TF Family: FAR1 |
FAR1-related sequence 5 [Theobroma cacao] |
| 17 |
Hb_009742_010 |
0.3164863625 |
- |
- |
- |
| 18 |
Hb_004939_020 |
0.3171409259 |
- |
- |
gag/pol protein [Bryonia dioica] |
| 19 |
Hb_092043_020 |
0.3255411217 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 20 |
Hb_008727_070 |
0.3284944355 |
- |
- |
PREDICTED: 16.9 kDa class I heat shock protein 1 isoform X1 [Zea mays] |