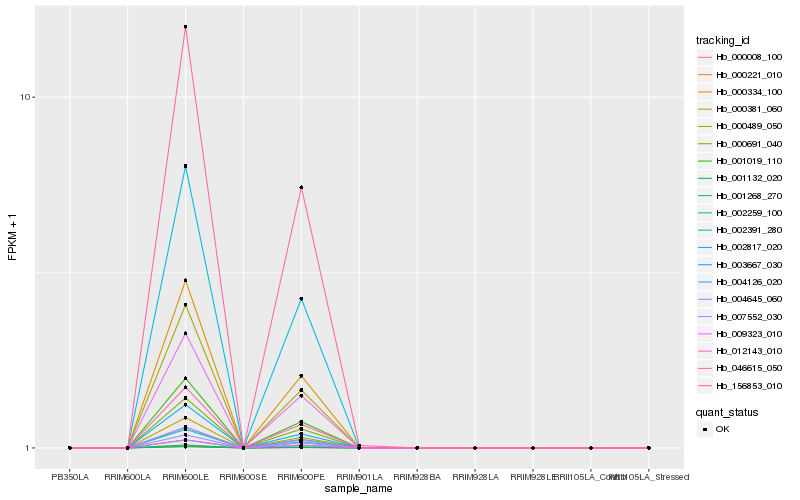
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_046615_050 |
0.0 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 2 |
Hb_000691_040 |
0.0009514337 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 3 |
Hb_001268_270 |
0.0014024205 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_012143_010 |
0.00335018 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 [Vitis vinifera] |
| 5 |
Hb_004126_020 |
0.0049890344 |
- |
- |
PREDICTED: uncharacterized protein LOC105649549 [Jatropha curcas] |
| 6 |
Hb_001132_020 |
0.0062666625 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 7 |
Hb_000381_060 |
0.0090939784 |
- |
- |
- |
| 8 |
Hb_001019_110 |
0.0095270182 |
- |
- |
PREDICTED: cytochrome P450 734A1 [Jatropha curcas] |
| 9 |
Hb_007552_030 |
0.0098057251 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21064 [Jatropha curcas] |
| 10 |
Hb_000221_010 |
0.0124804747 |
- |
- |
polyprotein [Oryza australiensis] |
| 11 |
Hb_009323_010 |
0.013146754 |
- |
- |
hypothetical protein POPTR_0008s03350g, partial [Populus trichocarpa] |
| 12 |
Hb_002817_020 |
0.0166835899 |
- |
- |
- |
| 13 |
Hb_002391_280 |
0.0170530403 |
- |
- |
PREDICTED: ammonium transporter 3 member 1-like [Jatropha curcas] |
| 14 |
Hb_000008_100 |
0.0181347539 |
- |
- |
Cyclic nucleotide-gated channel 15 [Theobroma cacao] |
| 15 |
Hb_000334_100 |
0.0217079539 |
- |
- |
hypothetical protein JCGZ_16788 [Jatropha curcas] |
| 16 |
Hb_004645_060 |
0.0245580028 |
- |
- |
PREDICTED: uncharacterized protein LOC105630948 [Jatropha curcas] |
| 17 |
Hb_000489_050 |
0.0252174141 |
- |
- |
PREDICTED: uncharacterized protein LOC105643984 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003667_030 |
0.0273081816 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL15 [Jatropha curcas] |
| 19 |
Hb_156853_010 |
0.0287578083 |
- |
- |
PREDICTED: vinorine synthase-like [Populus euphratica] |
| 20 |
Hb_002259_100 |
0.0294746439 |
- |
- |
PREDICTED: uncharacterized protein LOC105630152 isoform X1 [Jatropha curcas] |