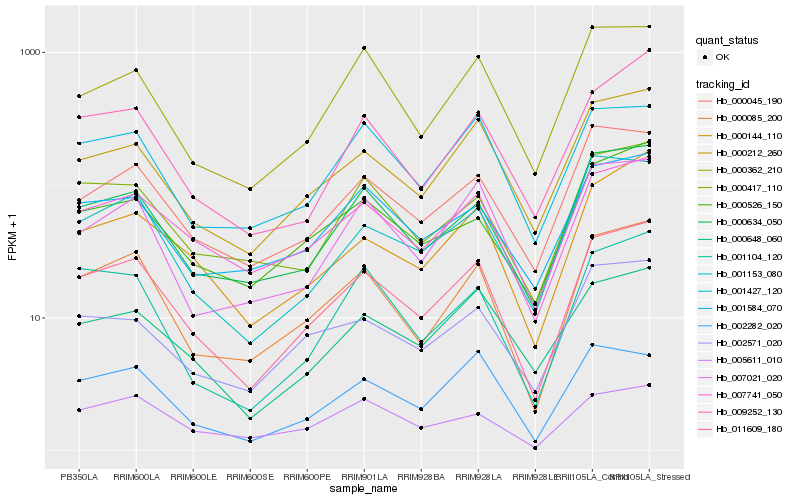
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011609_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0275933-like [Citrus sinensis] |
| 2 |
Hb_000085_200 |
0.0651630495 |
- |
- |
ATP synthase subunit H protein [Hevea brasiliensis] |
| 3 |
Hb_000634_050 |
0.082915265 |
- |
- |
PREDICTED: uncharacterized protein LOC105636623 [Jatropha curcas] |
| 4 |
Hb_000212_260 |
0.0853147499 |
- |
- |
acyl-carrier-protein [Triadica sebifera] |
| 5 |
Hb_007741_050 |
0.087435479 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 6 |
Hb_000144_110 |
0.0941699518 |
- |
- |
hypothetical protein CICLE_v10013189mg [Citrus clementina] |
| 7 |
Hb_000648_060 |
0.0977520207 |
- |
- |
hypothetical protein POPTR_0019s10570g [Populus trichocarpa] |
| 8 |
Hb_000045_190 |
0.0980001326 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit F [Jatropha curcas] |
| 9 |
Hb_001153_080 |
0.0992936692 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL19 [Jatropha curcas] |
| 10 |
Hb_000417_110 |
0.1002646611 |
- |
- |
PREDICTED: uncharacterized protein LOC105649681 [Jatropha curcas] |
| 11 |
Hb_000526_150 |
0.1029740581 |
- |
- |
hypothetical protein POPTR_0019s10790g, partial [Populus trichocarpa] |
| 12 |
Hb_007021_020 |
0.1039385706 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_009252_130 |
0.1049187145 |
- |
- |
Deletion of SUV3 suppressor 1(I) isoform 1 [Theobroma cacao] |
| 14 |
Hb_001104_120 |
0.1052296001 |
- |
- |
- |
| 15 |
Hb_001584_070 |
0.1053340305 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002282_020 |
0.1054492169 |
- |
- |
PREDICTED: uncharacterized protein LOC105629828 [Jatropha curcas] |
| 17 |
Hb_005611_010 |
0.1061256001 |
- |
- |
PREDICTED: protein prune homolog [Jatropha curcas] |
| 18 |
Hb_000362_210 |
0.1063541313 |
- |
- |
PREDICTED: transcription elongation factor 1 homolog [Jatropha curcas] |
| 19 |
Hb_002571_020 |
0.1064699298 |
- |
- |
PREDICTED: uncharacterized protein LOC105132457 isoform X12 [Populus euphratica] |
| 20 |
Hb_001427_120 |
0.1067813477 |
- |
- |
PREDICTED: uncharacterized protein LOC105637787 [Jatropha curcas] |