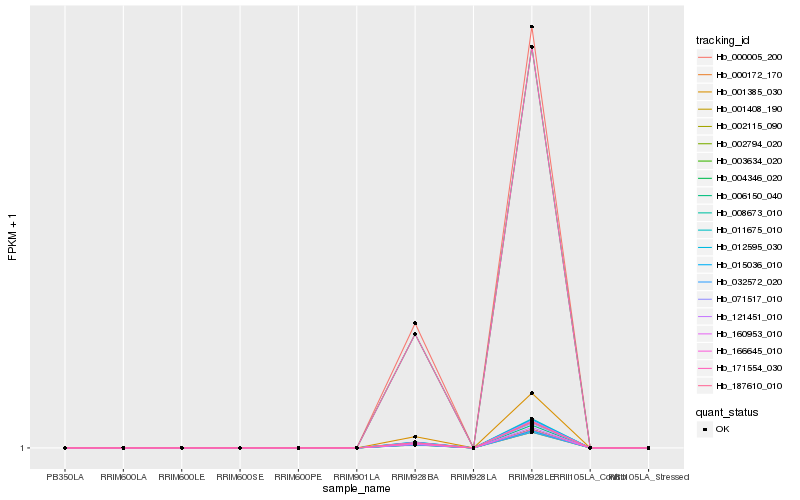
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006150_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105772114 [Gossypium raimondii] |
| 2 |
Hb_071517_010 |
0.0007751761 |
- |
- |
hypothetical protein CICLE_v10010288mg, partial [Citrus clementina] |
| 3 |
Hb_121451_010 |
0.0009369552 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 4 |
Hb_000005_200 |
0.001020614 |
- |
- |
- |
| 5 |
Hb_000172_170 |
0.0012175123 |
- |
- |
PREDICTED: uncharacterized protein LOC104908276 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_187610_010 |
0.0013729246 |
- |
- |
PREDICTED: uncharacterized protein LOC105785094 [Gossypium raimondii] |
| 7 |
Hb_002794_020 |
0.0016932015 |
- |
- |
PREDICTED: cytochrome P450 89A2-like [Jatropha curcas] |
| 8 |
Hb_003634_020 |
0.0017005876 |
- |
- |
PREDICTED: uncharacterized protein LOC105775392, partial [Gossypium raimondii] |
| 9 |
Hb_001408_190 |
0.0017393645 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 10 |
Hb_166645_010 |
0.0018002698 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105123834 [Populus euphratica] |
| 11 |
Hb_160953_010 |
0.0018028026 |
- |
- |
PREDICTED: uncharacterized protein LOC104214925 [Nicotiana sylvestris] |
| 12 |
Hb_008673_010 |
0.0020054607 |
- |
- |
PREDICTED: uncharacterized protein LOC104879858 [Vitis vinifera] |
| 13 |
Hb_012595_030 |
0.0023523375 |
- |
- |
- |
| 14 |
Hb_015036_010 |
0.0023843723 |
- |
- |
- |
| 15 |
Hb_001385_030 |
0.002384489 |
- |
- |
PREDICTED: uncharacterized protein LOC100852852 [Vitis vinifera] |
| 16 |
Hb_004346_020 |
0.0026884368 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_171554_030 |
0.0026884368 |
- |
- |
- |
| 18 |
Hb_011675_010 |
0.0027406048 |
- |
- |
- |
| 19 |
Hb_032572_020 |
0.0028314922 |
- |
- |
- |
| 20 |
Hb_002115_090 |
0.0028751913 |
- |
- |
PREDICTED: uncharacterized protein LOC104590556 [Nelumbo nucifera] |