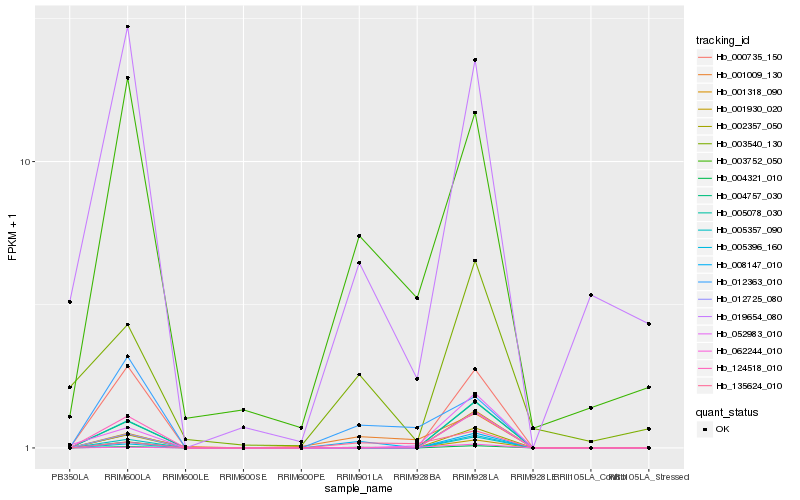
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004321_010 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_008147_010 |
0.1263360448 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 3 |
Hb_005078_030 |
0.2771666193 |
- |
- |
- |
| 4 |
Hb_004757_030 |
0.2772539827 |
- |
- |
- |
| 5 |
Hb_002357_050 |
0.2772611688 |
- |
- |
- |
| 6 |
Hb_001930_020 |
0.2811016682 |
- |
- |
PREDICTED: pectinesterase QRT1 [Jatropha curcas] |
| 7 |
Hb_005357_090 |
0.2819027751 |
- |
- |
ubiquitin, putative [Ricinus communis] |
| 8 |
Hb_135624_010 |
0.2832928408 |
- |
- |
PREDICTED: inositol transporter 4-like [Jatropha curcas] |
| 9 |
Hb_124518_010 |
0.2882662148 |
- |
- |
PREDICTED: uncharacterized protein LOC105763691 [Gossypium raimondii] |
| 10 |
Hb_003752_050 |
0.2886970437 |
- |
- |
hypothetical protein JCGZ_15078 [Jatropha curcas] |
| 11 |
Hb_001318_090 |
0.2918142853 |
- |
- |
PREDICTED: zinc finger protein 598 [Jatropha curcas] |
| 12 |
Hb_012725_080 |
0.2922515569 |
- |
- |
- |
| 13 |
Hb_001009_130 |
0.2947734205 |
- |
- |
hypothetical protein JCGZ_11336 [Jatropha curcas] |
| 14 |
Hb_003540_130 |
0.2976188059 |
- |
- |
- |
| 15 |
Hb_000735_150 |
0.3010367002 |
- |
- |
hypothetical protein JCGZ_15322 [Jatropha curcas] |
| 16 |
Hb_005396_160 |
0.3013443625 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 17 |
Hb_019654_080 |
0.3152113467 |
- |
- |
- |
| 18 |
Hb_012363_010 |
0.3271944703 |
- |
- |
- |
| 19 |
Hb_062244_010 |
0.3340081704 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_052983_010 |
0.3356218672 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |