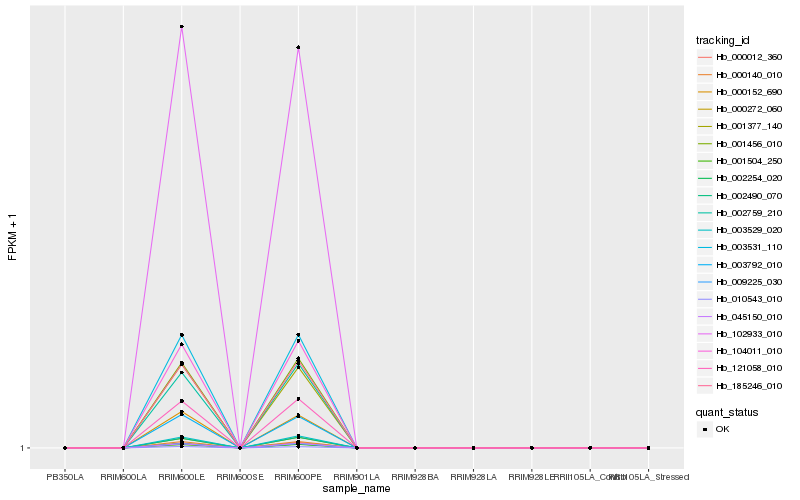
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003531_110 |
0.0 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 2 |
Hb_000140_010 |
0.0013392483 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 3 |
Hb_009225_030 |
0.0073921302 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 4 |
Hb_104011_010 |
0.008014181 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Jatropha curcas] |
| 5 |
Hb_001377_140 |
0.0090343009 |
- |
- |
hypothetical protein MIMGU_mgv1a016697mg [Erythranthe guttata] |
| 6 |
Hb_185246_010 |
0.0099138193 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 7 |
Hb_121058_010 |
0.0099692993 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_002254_020 |
0.0115055482 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 2 protein-like [Jatropha curcas] |
| 9 |
Hb_001456_010 |
0.0116333605 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_001504_250 |
0.0130296223 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_003792_010 |
0.0132538675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_045150_010 |
0.0184230577 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 13 |
Hb_102933_010 |
0.0187375399 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |
| 14 |
Hb_003529_020 |
0.0212857703 |
- |
- |
hypothetical protein JCGZ_04979 [Jatropha curcas] |
| 15 |
Hb_002490_070 |
0.0224685954 |
- |
- |
PREDICTED: uncharacterized protein LOC105123384 isoform X1 [Populus euphratica] |
| 16 |
Hb_000272_060 |
0.0233612494 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_000152_690 |
0.0239073336 |
- |
- |
PREDICTED: uncharacterized protein LOC105648869 [Jatropha curcas] |
| 18 |
Hb_000012_360 |
0.0242833498 |
- |
- |
PREDICTED: uncharacterized protein LOC104248159 [Nicotiana sylvestris] |
| 19 |
Hb_002759_210 |
0.0261147428 |
- |
- |
Peroxidase 6 precursor family protein [Populus trichocarpa] |
| 20 |
Hb_010543_010 |
0.0266584389 |
- |
- |
Phospholipid-transporting ATPase, putative [Ricinus communis] |