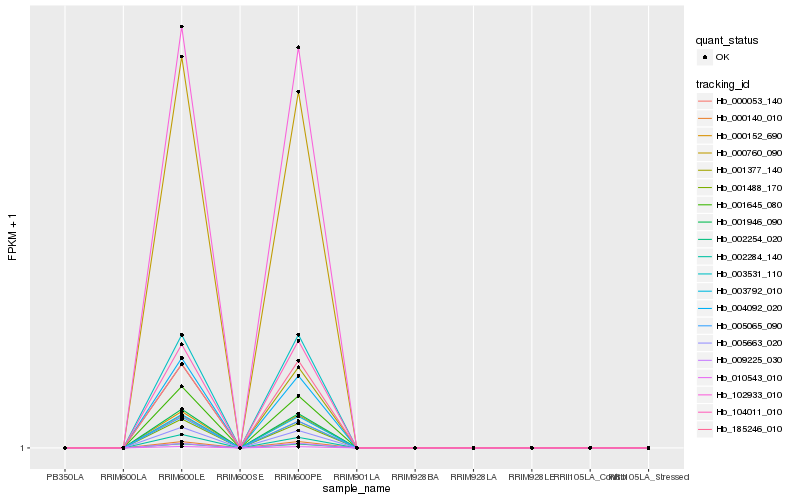
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_690 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648869 [Jatropha curcas] |
| 2 |
Hb_010543_010 |
0.002751715 |
- |
- |
Phospholipid-transporting ATPase, putative [Ricinus communis] |
| 3 |
Hb_001946_090 |
0.0037946737 |
- |
- |
- |
| 4 |
Hb_102933_010 |
0.0051705981 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |
| 5 |
Hb_005065_090 |
0.0088602626 |
- |
- |
zinc/iron transporter, putative [Ricinus communis] |
| 6 |
Hb_000760_090 |
0.009101539 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-6 [Vitis vinifera] |
| 7 |
Hb_003792_010 |
0.010654638 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002254_020 |
0.0124029695 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 2 protein-like [Jatropha curcas] |
| 9 |
Hb_001488_170 |
0.0125540355 |
- |
- |
PREDICTED: transcription initiation factor IIB-like [Jatropha curcas] |
| 10 |
Hb_001645_080 |
0.0145521824 |
transcription factor |
TF Family: C2H2 |
palmate-like pentafoliata 1 transcription factor [Manihot esculenta] |
| 11 |
Hb_001377_140 |
0.0148741475 |
- |
- |
hypothetical protein MIMGU_mgv1a016697mg [Erythranthe guttata] |
| 12 |
Hb_005663_020 |
0.0150806678 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 13 |
Hb_000053_140 |
0.0191566573 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003531_110 |
0.0239073336 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 15 |
Hb_000140_010 |
0.0252463015 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 16 |
Hb_004092_020 |
0.0287310612 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 32 [Jatropha curcas] |
| 17 |
Hb_002284_140 |
0.029128048 |
- |
- |
hypothetical protein POPTR_0002s00800g [Populus trichocarpa] |
| 18 |
Hb_009225_030 |
0.0312975462 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 19 |
Hb_104011_010 |
0.0319193944 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Jatropha curcas] |
| 20 |
Hb_185246_010 |
0.0338183743 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |