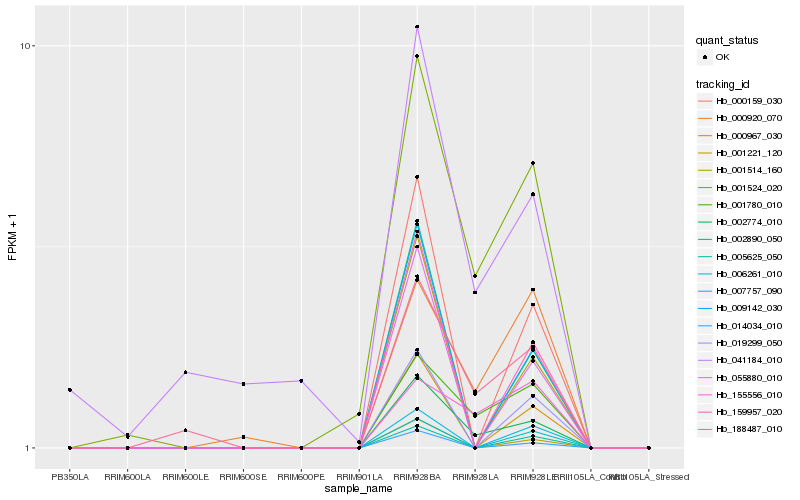
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002774_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_001524_020 |
0.1259568361 |
- |
- |
Pollen-specific protein C13 precursor, putative [Ricinus communis] |
| 3 |
Hb_001780_010 |
0.1341294855 |
- |
- |
hypothetical protein CICLE_v10006597mg [Citrus clementina] |
| 4 |
Hb_188487_010 |
0.1549218409 |
- |
- |
hypothetical protein JCGZ_24382 [Jatropha curcas] |
| 5 |
Hb_000920_070 |
0.2158245006 |
- |
- |
- |
| 6 |
Hb_155556_010 |
0.2233115763 |
- |
- |
putative methylenetetrahydrofolate reductase [Morus notabilis] |
| 7 |
Hb_159957_020 |
0.2277363139 |
- |
- |
- |
| 8 |
Hb_000159_030 |
0.2277859007 |
- |
- |
putative glycosyltransferase, partial [Aralia elata] |
| 9 |
Hb_055880_010 |
0.2282808691 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_009142_030 |
0.2285156473 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_001514_160 |
0.228687296 |
- |
- |
hypothetical protein JCGZ_16070 [Jatropha curcas] |
| 12 |
Hb_000967_030 |
0.2290063039 |
- |
- |
- |
| 13 |
Hb_014034_010 |
0.2291662814 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_002890_050 |
0.229276515 |
- |
- |
- |
| 15 |
Hb_001221_120 |
0.2295732164 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_019299_050 |
0.2363603227 |
- |
- |
- |
| 17 |
Hb_005625_050 |
0.2429076586 |
- |
- |
hypothetical protein PRUPE_ppa024499mg, partial [Prunus persica] |
| 18 |
Hb_007757_090 |
0.2456422311 |
- |
- |
PREDICTED: uncharacterized protein LOC105793478 [Gossypium raimondii] |
| 19 |
Hb_041184_010 |
0.2459611302 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 20 |
Hb_006261_010 |
0.2480399551 |
- |
- |
PREDICTED: uncharacterized protein LOC104110884 [Nicotiana tomentosiformis] |