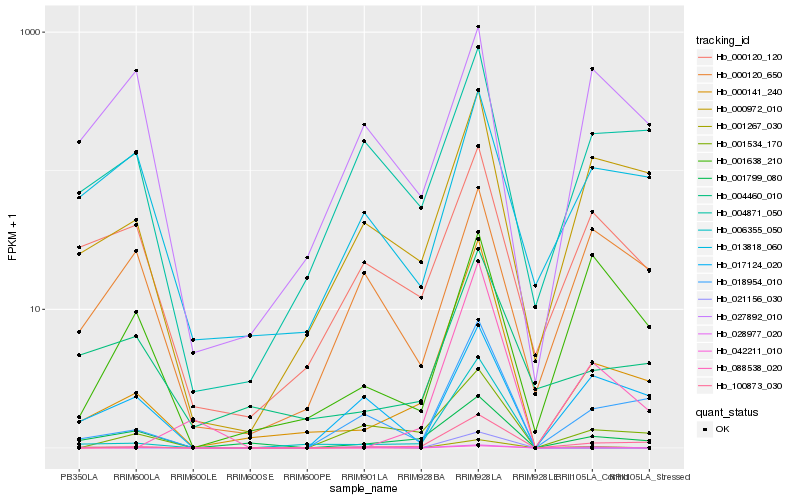
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001267_030 |
0.0 |
- |
- |
hypothetical protein VITISV_041979 [Vitis vinifera] |
| 2 |
Hb_000141_240 |
0.268519833 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_018954_010 |
0.2792842959 |
- |
- |
- |
| 4 |
Hb_017124_020 |
0.2814087914 |
- |
- |
hypothetical protein PRUPE_ppa015539mg [Prunus persica] |
| 5 |
Hb_028977_020 |
0.2860486258 |
- |
- |
hypothetical protein JCGZ_21319 [Jatropha curcas] |
| 6 |
Hb_000120_120 |
0.2972410871 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 2 [Jatropha curcas] |
| 7 |
Hb_001799_080 |
0.3106665928 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 8 |
Hb_000972_010 |
0.317684609 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 9 |
Hb_001534_170 |
0.3194106322 |
- |
- |
Uncharacterized protein TCM_038456 [Theobroma cacao] |
| 10 |
Hb_021156_030 |
0.3206265112 |
- |
- |
hypothetical protein JCGZ_18422 [Jatropha curcas] |
| 11 |
Hb_100873_030 |
0.3283681438 |
- |
- |
PREDICTED: uncharacterized protein LOC105795653 [Gossypium raimondii] |
| 12 |
Hb_006355_050 |
0.3288110669 |
- |
- |
- |
| 13 |
Hb_004871_050 |
0.3302051006 |
- |
- |
hypothetical protein F383_22684 [Gossypium arboreum] |
| 14 |
Hb_004460_010 |
0.3365320326 |
- |
- |
PREDICTED: SKI/DACH domain-containing protein 1-like [Jatropha curcas] |
| 15 |
Hb_013818_060 |
0.3396430641 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 16 |
Hb_027892_010 |
0.3462271773 |
transcription factor |
TF Family: bHLH |
LMYC1 [Hevea brasiliensis] |
| 17 |
Hb_000120_650 |
0.3462318252 |
- |
- |
PREDICTED: proton-coupled amino acid transporter 3 [Jatropha curcas] |
| 18 |
Hb_088538_020 |
0.3471918103 |
- |
- |
hypothetical protein JCGZ_12178 [Jatropha curcas] |
| 19 |
Hb_001638_210 |
0.3536496697 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_042211_010 |
0.3570185241 |
- |
- |
PREDICTED: uncharacterized protein LOC103404019 [Malus domestica] |