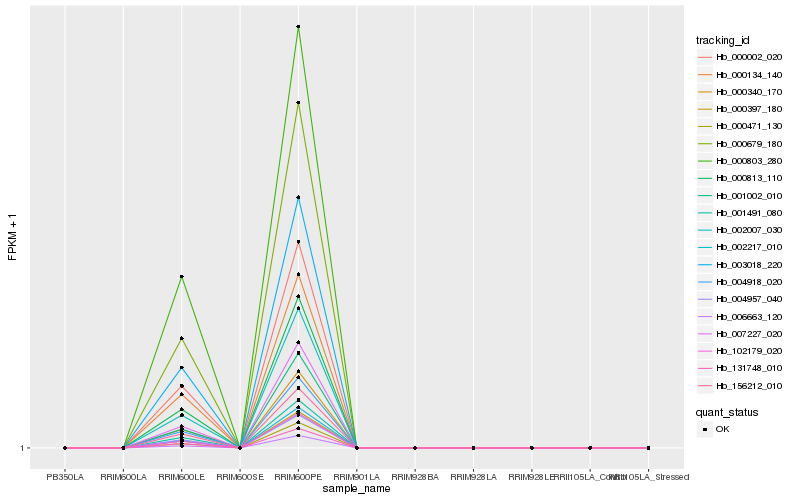
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000813_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104884346 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_000803_280 |
0.0057237894 |
- |
- |
PREDICTED: uncharacterized protein LOC105648326 [Jatropha curcas] |
| 3 |
Hb_001491_080 |
0.0065712929 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000397_180 |
0.0078926646 |
- |
- |
- |
| 5 |
Hb_102179_020 |
0.0092425837 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 6 |
Hb_003018_220 |
0.0101776704 |
- |
- |
Snakin-2 [Gossypium arboreum] |
| 7 |
Hb_004918_020 |
0.0105002179 |
- |
- |
- |
| 8 |
Hb_002007_030 |
0.0106545679 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000002_020 |
0.0121625624 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 10 |
Hb_156212_010 |
0.0129191438 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 11 |
Hb_000340_170 |
0.0130040822 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g54790-like [Jatropha curcas] |
| 12 |
Hb_131748_010 |
0.0188016936 |
- |
- |
hypothetical protein PRUPE_ppa022115mg [Prunus persica] |
| 13 |
Hb_007227_020 |
0.0216245413 |
- |
- |
- |
| 14 |
Hb_000679_180 |
0.0220435776 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_006663_120 |
0.0223405979 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_001002_010 |
0.0235443142 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000134_140 |
0.0270476763 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 18 |
Hb_002217_010 |
0.0284631045 |
- |
- |
Peroxiredoxin, putative [Ricinus communis] |
| 19 |
Hb_000471_130 |
0.0293566186 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004957_040 |
0.0303741007 |
- |
- |
PREDICTED: uncharacterized protein LOC104890338 [Beta vulgaris subsp. vulgaris] |