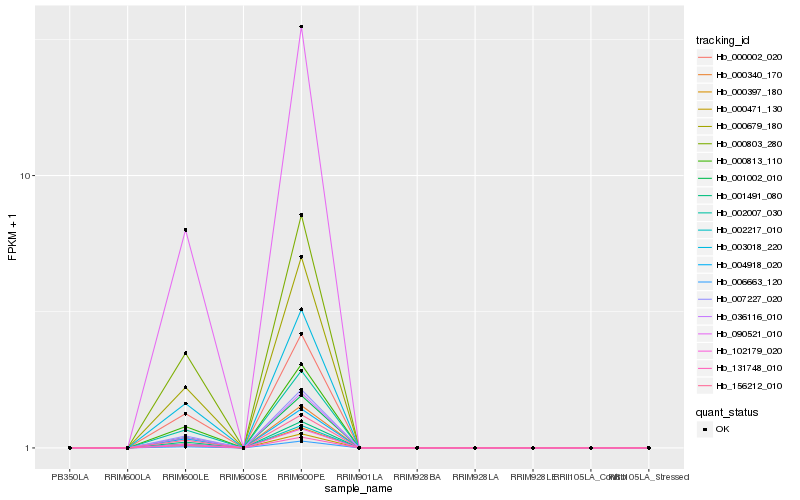
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000397_180 |
0.0 |
- |
- |
- |
| 2 |
Hb_102179_020 |
0.0013500145 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 3 |
Hb_002007_030 |
0.0027621288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000813_110 |
0.0078926646 |
- |
- |
PREDICTED: uncharacterized protein LOC104884346 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_000803_280 |
0.0136158761 |
- |
- |
PREDICTED: uncharacterized protein LOC105648326 [Jatropha curcas] |
| 6 |
Hb_007227_020 |
0.0137342039 |
- |
- |
- |
| 7 |
Hb_000679_180 |
0.0141533597 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_006663_120 |
0.0144504666 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_001491_080 |
0.0144632539 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001002_010 |
0.015654549 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003018_220 |
0.0180689833 |
- |
- |
Snakin-2 [Gossypium arboreum] |
| 12 |
Hb_004918_020 |
0.0183914639 |
- |
- |
- |
| 13 |
Hb_000002_020 |
0.0200534413 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 14 |
Hb_002217_010 |
0.0205750933 |
- |
- |
Peroxiredoxin, putative [Ricinus communis] |
| 15 |
Hb_156212_010 |
0.020809843 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 16 |
Hb_000340_170 |
0.0208947607 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g54790-like [Jatropha curcas] |
| 17 |
Hb_000471_130 |
0.0214689711 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_090521_010 |
0.0239460956 |
- |
- |
Major allergen Pru ar, putative [Ricinus communis] |
| 19 |
Hb_036116_010 |
0.0256557413 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 20 |
Hb_131748_010 |
0.0266907319 |
- |
- |
hypothetical protein PRUPE_ppa022115mg [Prunus persica] |