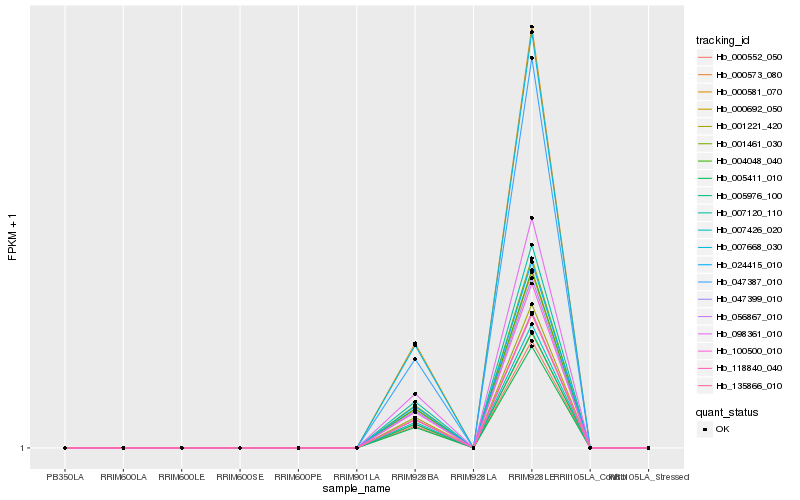
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000573_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC101510272 [Cicer arietinum] |
| 2 |
Hb_004048_040 |
0.0003459328 |
- |
- |
- |
| 3 |
Hb_047399_010 |
0.0008423013 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH2 [Manihot esculenta] |
| 4 |
Hb_024415_010 |
0.0015644372 |
- |
- |
- |
| 5 |
Hb_100500_010 |
0.0016230122 |
- |
- |
PREDICTED: uncharacterized protein LOC105785446 [Gossypium raimondii] |
| 6 |
Hb_001461_030 |
0.0020781732 |
- |
- |
- |
| 7 |
Hb_007120_110 |
0.0041712931 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 8 |
Hb_001221_420 |
0.0045222678 |
- |
- |
Uncharacterized protein TCM_032186 [Theobroma cacao] |
| 9 |
Hb_047387_010 |
0.0046141223 |
- |
- |
- |
| 10 |
Hb_056867_010 |
0.0056416295 |
- |
- |
PREDICTED: uncharacterized protein LOC105786773 [Gossypium raimondii] |
| 11 |
Hb_118840_040 |
0.0057220132 |
- |
- |
- |
| 12 |
Hb_135866_010 |
0.0058807583 |
- |
- |
PREDICTED: uncharacterized protein LOC105778857 [Gossypium raimondii] |
| 13 |
Hb_007426_020 |
0.0072416824 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 14 |
Hb_007668_030 |
0.0074264827 |
- |
- |
PREDICTED: cold shock domain-containing protein 3-like [Musa acuminata subsp. malaccensis] |
| 15 |
Hb_000581_070 |
0.0079590055 |
- |
- |
- |
| 16 |
Hb_005976_100 |
0.008239582 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein RCOM_1408460 [Ricinus communis] |
| 17 |
Hb_000692_050 |
0.0083520345 |
- |
- |
- |
| 18 |
Hb_098361_010 |
0.00841778 |
- |
- |
PREDICTED: uncharacterized protein LOC105118398 [Populus euphratica] |
| 19 |
Hb_000552_050 |
0.009387315 |
- |
- |
- |
| 20 |
Hb_005411_010 |
0.0101076754 |
- |
- |
- |