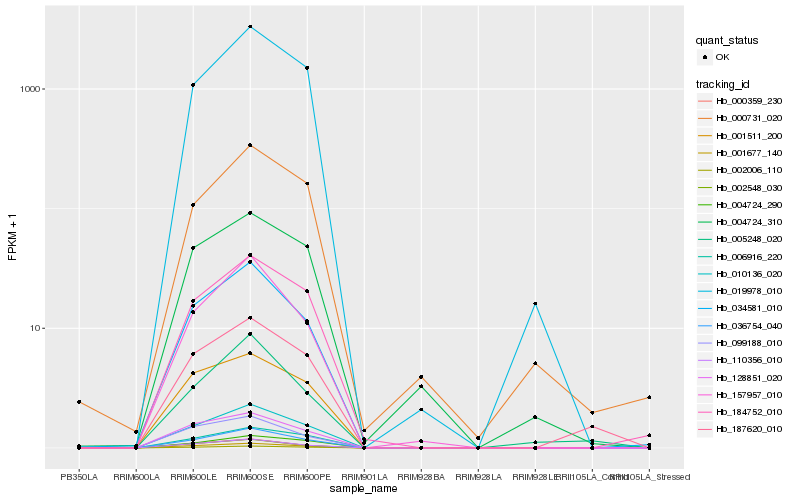
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_187620_010 |
0.0 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_010136_020 |
0.1110116176 |
- |
- |
stress-induced receptor-like kinase [Medicago truncatula] |
| 3 |
Hb_002006_110 |
0.1112475037 |
transcription factor |
TF Family: LFY |
LEAFY-like protein [Pistacia chinensis] |
| 4 |
Hb_001677_140 |
0.1134090458 |
- |
- |
PREDICTED: uncharacterized protein LOC102622520 isoform X1 [Citrus sinensis] |
| 5 |
Hb_036754_040 |
0.1167123627 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 6 |
Hb_001511_200 |
0.122157055 |
- |
- |
Embryonic abundant protein, putative [Ricinus communis] |
| 7 |
Hb_128851_020 |
0.124807727 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_184752_010 |
0.1259332201 |
- |
- |
PREDICTED: uncharacterized protein LOC105642593 [Jatropha curcas] |
| 9 |
Hb_019978_010 |
0.1285862625 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_034581_010 |
0.1292473055 |
- |
- |
PREDICTED: acidic mammalian chitinase-like [Jatropha curcas] |
| 11 |
Hb_004724_290 |
0.1293890826 |
transcription factor |
TF Family: NAC |
- |
| 12 |
Hb_110356_010 |
0.1313325423 |
- |
- |
hypothetical protein JCGZ_21614 [Jatropha curcas] |
| 13 |
Hb_006916_220 |
0.1351630559 |
- |
- |
hypothetical protein JCGZ_20099 [Jatropha curcas] |
| 14 |
Hb_005248_020 |
0.1389624173 |
- |
- |
hypothetical protein JCGZ_23770 [Jatropha curcas] |
| 15 |
Hb_002548_030 |
0.1398979648 |
- |
- |
PREDICTED: uncharacterized protein LOC103343650 [Prunus mume] |
| 16 |
Hb_004724_310 |
0.140899285 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 17 |
Hb_000359_230 |
0.1411377915 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Prunus mume] |
| 18 |
Hb_099188_010 |
0.1443097504 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 19 |
Hb_000731_020 |
0.1475230308 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 20 |
Hb_157957_010 |
0.1501674203 |
- |
- |
hypothetical protein JCGZ_10722 [Jatropha curcas] |