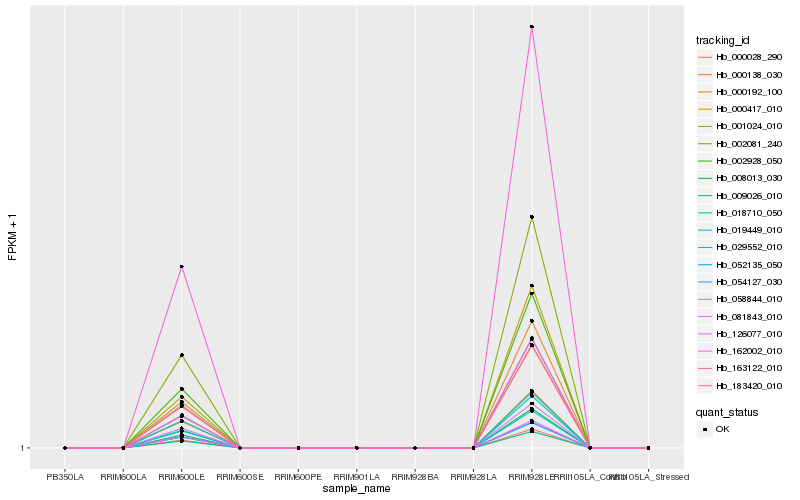
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_163122_010 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_002928_050 |
0.013298527 |
- |
- |
- |
| 3 |
Hb_002081_240 |
0.0168565279 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 4 |
Hb_018710_050 |
0.0177186257 |
- |
- |
- |
| 5 |
Hb_008013_030 |
0.0210262005 |
- |
- |
- |
| 6 |
Hb_126077_010 |
0.025949624 |
- |
- |
- |
| 7 |
Hb_001024_010 |
0.0271053298 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 8 |
Hb_000028_290 |
0.0271836769 |
- |
- |
hypothetical protein JCGZ_25310 [Jatropha curcas] |
| 9 |
Hb_052135_050 |
0.0304495161 |
- |
- |
PREDICTED: probable pectinesterase 67 [Jatropha curcas] |
| 10 |
Hb_009026_010 |
0.0311242935 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 11 |
Hb_081843_010 |
0.0346000332 |
- |
- |
PREDICTED: uncharacterized protein LOC104880938 [Vitis vinifera] |
| 12 |
Hb_054127_030 |
0.0357098097 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase II.2 [Jatropha curcas] |
| 13 |
Hb_162002_010 |
0.0372820585 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 14 |
Hb_000138_030 |
0.0391054529 |
- |
- |
- |
| 15 |
Hb_000192_100 |
0.041776608 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 16 |
Hb_019449_010 |
0.0422118408 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_183420_010 |
0.0432418108 |
- |
- |
PREDICTED: uncharacterized protein LOC105781565 [Gossypium raimondii] |
| 18 |
Hb_000417_010 |
0.0518966196 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase-like [Jatropha curcas] |
| 19 |
Hb_058844_010 |
0.0522367492 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 20 |
Hb_029552_010 |
0.0533932581 |
- |
- |
hypothetical protein JCGZ_08684 [Jatropha curcas] |