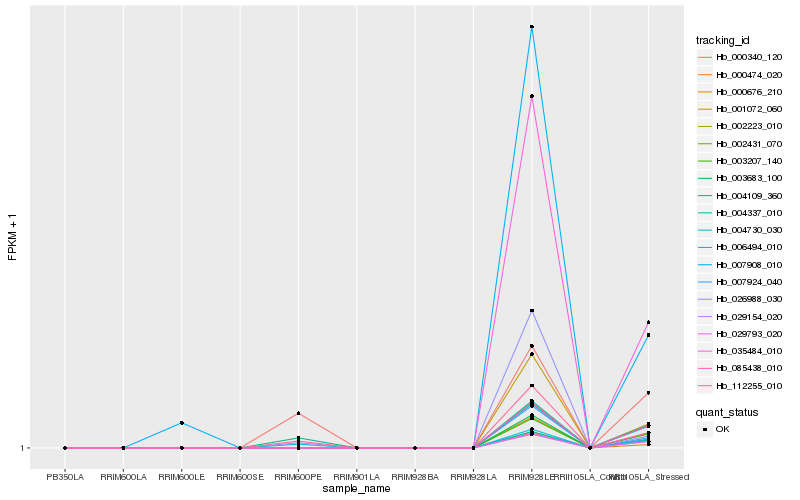
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_112255_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 2 |
Hb_006494_010 |
0.1314066244 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 3 |
Hb_004109_360 |
0.1391016127 |
- |
- |
PREDICTED: omega-6 fatty acid desaturase, endoplasmic reticulum isozyme 1-like [Jatropha curcas] |
| 4 |
Hb_026988_030 |
0.1735593018 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 5 |
Hb_000340_120 |
0.1840464132 |
- |
- |
PREDICTED: uncharacterized protein LOC103406091 [Malus domestica] |
| 6 |
Hb_007924_040 |
0.2006491033 |
- |
- |
PREDICTED: pollen-specific leucine-rich repeat extensin-like protein 3 [Populus euphratica] |
| 7 |
Hb_003683_100 |
0.200659136 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 8 |
Hb_001072_060 |
0.2006645245 |
- |
- |
PREDICTED: uncharacterized protein LOC105645525 [Jatropha curcas] |
| 9 |
Hb_000474_020 |
0.2006881845 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 10 |
Hb_035484_010 |
0.2042846355 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1-like [Jatropha curcas] |
| 11 |
Hb_029154_020 |
0.2340307062 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_029793_020 |
0.2340922072 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_002223_010 |
0.234298464 |
- |
- |
hypothetical protein VITISV_009489 [Vitis vinifera] |
| 14 |
Hb_004337_010 |
0.2343058774 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 15 |
Hb_003207_140 |
0.2357629874 |
- |
- |
PREDICTED: protein HOTHEAD [Jatropha curcas] |
| 16 |
Hb_000676_210 |
0.2359123278 |
- |
- |
Cytochrome P450 superfamily protein [Theobroma cacao] |
| 17 |
Hb_004730_030 |
0.237472993 |
- |
- |
synaptonemal complex protein, putative [Ricinus communis] |
| 18 |
Hb_002431_070 |
0.237611296 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA11 [Jatropha curcas] |
| 19 |
Hb_085438_010 |
0.2376592454 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 20 |
Hb_007908_010 |
0.2395028249 |
- |
- |
- |