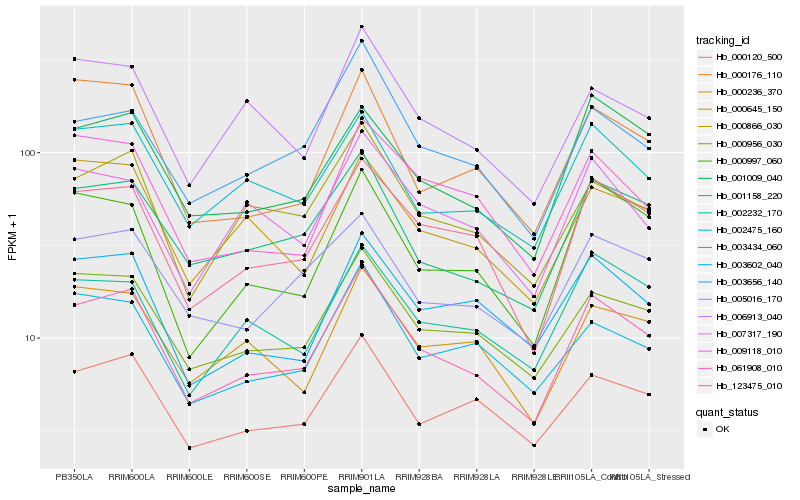
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_061908_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_16890 [Jatropha curcas] |
| 2 |
Hb_123475_010 |
0.0652492354 |
- |
- |
PREDICTED: hepatocyte growth factor-regulated tyrosine kinase substrate [Jatropha curcas] |
| 3 |
Hb_000956_030 |
0.0691169946 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 19-like [Jatropha curcas] |
| 4 |
Hb_001158_220 |
0.0732360965 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 5 |
Hb_000120_500 |
0.0792638193 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 10-like [Jatropha curcas] |
| 6 |
Hb_002475_160 |
0.0805024257 |
- |
- |
PREDICTED: 60S ribosomal export protein NMD3 [Jatropha curcas] |
| 7 |
Hb_001009_040 |
0.08429977 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 1 [Jatropha curcas] |
| 8 |
Hb_000866_030 |
0.0845879699 |
- |
- |
hypothetical protein RCOM_1615820 [Ricinus communis] |
| 9 |
Hb_000997_060 |
0.0862696869 |
- |
- |
PREDICTED: DNA repair protein XRCC4 [Jatropha curcas] |
| 10 |
Hb_005016_170 |
0.0863524024 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003602_040 |
0.0890298092 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 12 |
Hb_000645_150 |
0.0910093958 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_000236_370 |
0.091365619 |
- |
- |
PREDICTED: DNA-directed RNA polymerases I and III subunit RPAC1 [Jatropha curcas] |
| 14 |
Hb_009118_010 |
0.0921146011 |
- |
- |
methionine aminopeptidase, putative [Ricinus communis] |
| 15 |
Hb_003434_060 |
0.0925005286 |
- |
- |
PREDICTED: membrane-bound transcription factor site-2 protease homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_002232_170 |
0.0926380542 |
- |
- |
PREDICTED: uncharacterized protein LOC105635984 [Jatropha curcas] |
| 17 |
Hb_000176_110 |
0.0932692102 |
- |
- |
PREDICTED: serine--tRNA ligase [Jatropha curcas] |
| 18 |
Hb_006913_040 |
0.0947144669 |
- |
- |
PREDICTED: hydroxyacylglutathione hydrolase cytoplasmic [Jatropha curcas] |
| 19 |
Hb_003656_140 |
0.0952985776 |
- |
- |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Jatropha curcas] |
| 20 |
Hb_007317_190 |
0.0962058333 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI1 [Jatropha curcas] |