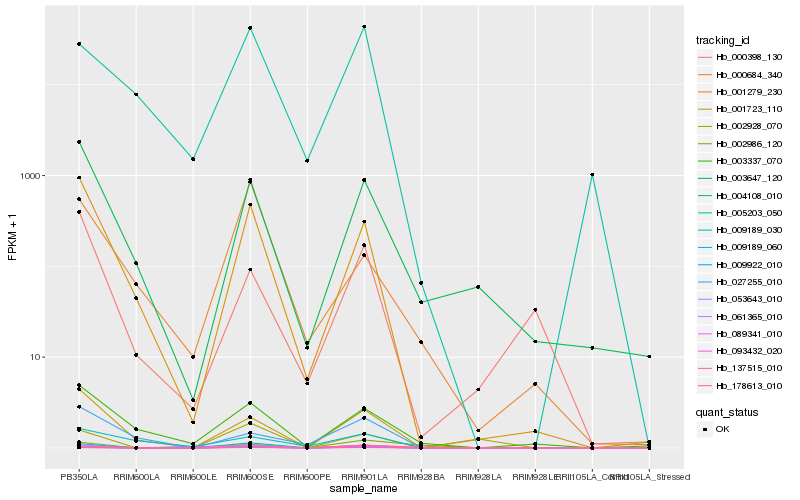
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_053643_010 |
0.0 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 2 |
Hb_009922_010 |
0.044566849 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 3 |
Hb_093432_020 |
0.0971443186 |
- |
- |
PREDICTED: QWRF motif-containing protein 3 [Jatropha curcas] |
| 4 |
Hb_002928_070 |
0.0998027976 |
- |
- |
- |
| 5 |
Hb_061365_010 |
0.1424345143 |
- |
- |
PREDICTED: uncharacterized protein LOC104227769, partial [Nicotiana sylvestris] |
| 6 |
Hb_137515_010 |
0.1719055746 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X1 [Populus euphratica] |
| 7 |
Hb_001279_230 |
0.2169806202 |
- |
- |
PREDICTED: class I heat shock protein-like [Jatropha curcas] |
| 8 |
Hb_005203_050 |
0.2387630572 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 9 |
Hb_003647_120 |
0.2587317491 |
- |
- |
18.2 kDa class I heat shock family protein [Populus trichocarpa] |
| 10 |
Hb_003337_070 |
0.2819706008 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 11 |
Hb_001723_110 |
0.2899606175 |
- |
- |
PREDICTED: solute carrier family 25 member 44-like isoform X2 [Malus domestica] |
| 12 |
Hb_004108_010 |
0.2935151295 |
- |
- |
- |
| 13 |
Hb_089341_010 |
0.3036029027 |
- |
- |
Uncharacterized protein TCM_018339 [Theobroma cacao] |
| 14 |
Hb_000684_340 |
0.3053561308 |
- |
- |
PREDICTED: L-ascorbate peroxidase 2, cytosolic [Pyrus x bretschneideri] |
| 15 |
Hb_002986_120 |
0.3109826428 |
- |
- |
- |
| 16 |
Hb_000398_130 |
0.3149018939 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein-like [Jatropha curcas] |
| 17 |
Hb_009189_060 |
0.3157300986 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g49170, chloroplastic [Jatropha curcas] |
| 18 |
Hb_009189_030 |
0.3193450965 |
- |
- |
hypothetical protein PRUPE_ppa023969mg, partial [Prunus persica] |
| 19 |
Hb_178613_010 |
0.3196476278 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 20 |
Hb_027255_010 |
0.3203717802 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |