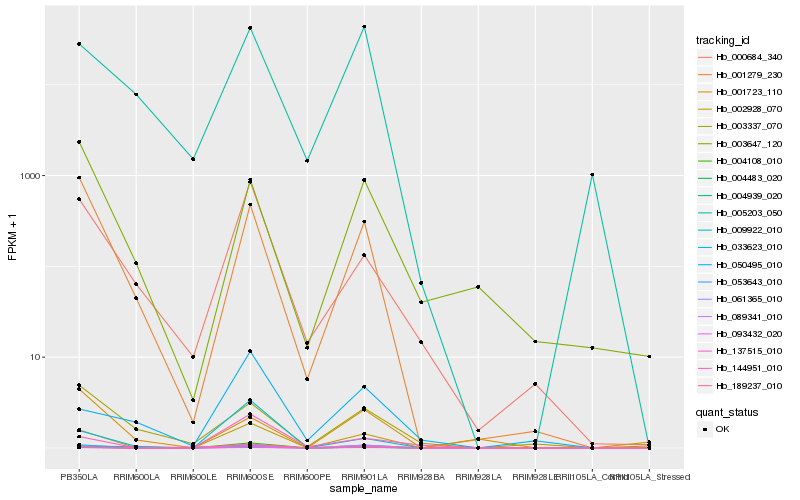
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009922_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 2 |
Hb_053643_010 |
0.044566849 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 3 |
Hb_093432_020 |
0.0541265572 |
- |
- |
PREDICTED: QWRF motif-containing protein 3 [Jatropha curcas] |
| 4 |
Hb_002928_070 |
0.0623760303 |
- |
- |
- |
| 5 |
Hb_061365_010 |
0.1654909138 |
- |
- |
PREDICTED: uncharacterized protein LOC104227769, partial [Nicotiana sylvestris] |
| 6 |
Hb_137515_010 |
0.191792492 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X1 [Populus euphratica] |
| 7 |
Hb_005203_050 |
0.2373695188 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 8 |
Hb_001279_230 |
0.237596331 |
- |
- |
PREDICTED: class I heat shock protein-like [Jatropha curcas] |
| 9 |
Hb_004108_010 |
0.2504968911 |
- |
- |
- |
| 10 |
Hb_003647_120 |
0.2843898195 |
- |
- |
18.2 kDa class I heat shock family protein [Populus trichocarpa] |
| 11 |
Hb_000684_340 |
0.2856528629 |
- |
- |
PREDICTED: L-ascorbate peroxidase 2, cytosolic [Pyrus x bretschneideri] |
| 12 |
Hb_144951_010 |
0.2864264975 |
- |
- |
putative reverse transcriptase [Oryza sativa Japonica Group] |
| 13 |
Hb_003337_070 |
0.2971572732 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 14 |
Hb_089341_010 |
0.2993856398 |
- |
- |
Uncharacterized protein TCM_018339 [Theobroma cacao] |
| 15 |
Hb_033623_010 |
0.309244008 |
- |
- |
- |
| 16 |
Hb_001723_110 |
0.3163202146 |
- |
- |
PREDICTED: solute carrier family 25 member 44-like isoform X2 [Malus domestica] |
| 17 |
Hb_004483_020 |
0.3209529822 |
- |
- |
- |
| 18 |
Hb_004939_020 |
0.3213471694 |
- |
- |
gag/pol protein [Bryonia dioica] |
| 19 |
Hb_189237_010 |
0.323466405 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 20 |
Hb_050495_010 |
0.3244975845 |
- |
- |
heat-shock protein, putative [Ricinus communis] |