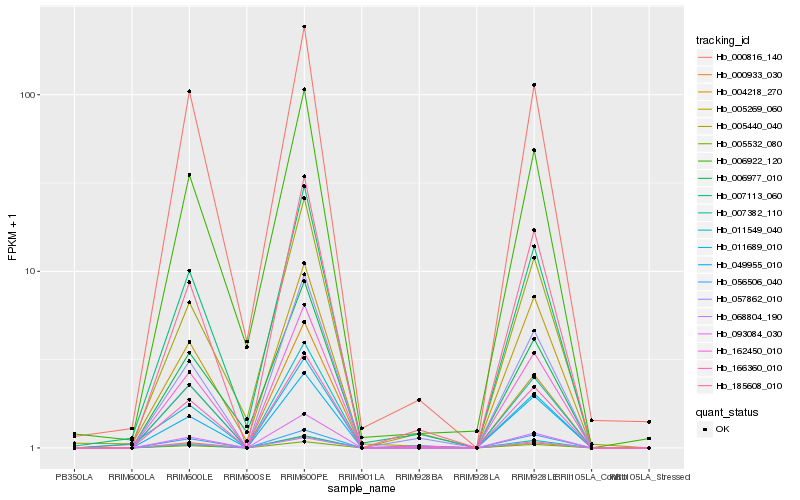
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011689_010 |
0.0 |
- |
- |
PREDICTED: S-adenosylmethionine decarboxylase proenzyme 4 [Jatropha curcas] |
| 2 |
Hb_166360_010 |
0.0828876448 |
- |
- |
hypothetical protein POPTR_0010s12130g [Populus trichocarpa] |
| 3 |
Hb_004218_270 |
0.0879013465 |
- |
- |
hypothetical protein CICLE_v100134152mg, partial [Citrus clementina] |
| 4 |
Hb_005532_080 |
0.0893106859 |
- |
- |
PREDICTED: heparanase-like protein 2 [Jatropha curcas] |
| 5 |
Hb_162450_010 |
0.0905639352 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 6 |
Hb_093084_030 |
0.0915268919 |
- |
- |
oleosin2 [Plukenetia volubilis] |
| 7 |
Hb_011549_040 |
0.0919288278 |
- |
- |
- |
| 8 |
Hb_068804_190 |
0.0938721757 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_007113_060 |
0.0942350195 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 10 |
Hb_000933_030 |
0.097472097 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007382_110 |
0.1081421243 |
- |
- |
PREDICTED: bidirectional sugar transporter N3 [Jatropha curcas] |
| 12 |
Hb_000816_140 |
0.1109394492 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 13 |
Hb_185608_010 |
0.1127137071 |
- |
- |
hypothetical protein POPTR_0004s22210g [Populus trichocarpa] |
| 14 |
Hb_049955_010 |
0.1165328729 |
- |
- |
hypothetical protein EUGRSUZ_B00742 [Eucalyptus grandis] |
| 15 |
Hb_057862_010 |
0.1171919965 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_056506_040 |
0.118043707 |
- |
- |
- |
| 17 |
Hb_005269_060 |
0.1204170736 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Cicer arietinum] |
| 18 |
Hb_006922_120 |
0.1216039668 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 19 |
Hb_005440_040 |
0.1235195718 |
- |
- |
PREDICTED: microtubule-associated protein 70-5 [Jatropha curcas] |
| 20 |
Hb_006977_010 |
0.1245673028 |
- |
- |
unnamed protein product [Vitis vinifera] |