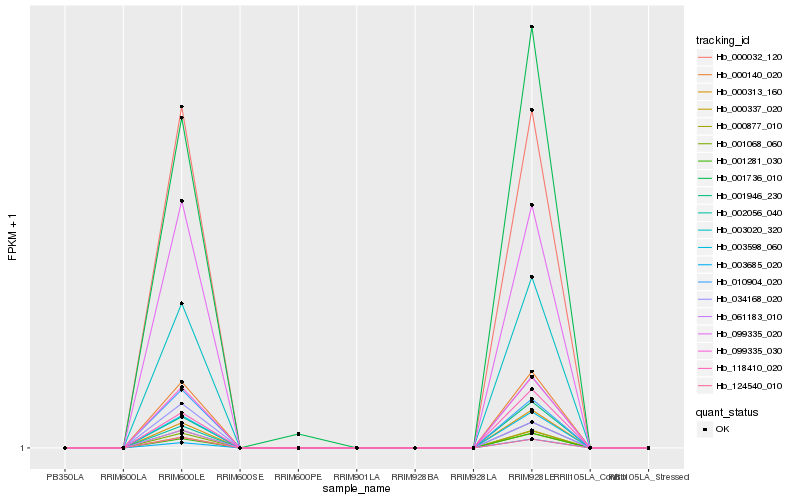
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003020_320 |
0.0 |
- |
- |
- |
| 2 |
Hb_099335_030 |
0.0121061552 |
- |
- |
hypothetical protein M569_10107, partial [Genlisea aurea] |
| 3 |
Hb_000140_020 |
0.0139569646 |
- |
- |
BnaA02g09930D [Brassica napus] |
| 4 |
Hb_002056_040 |
0.0144252095 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 5 |
Hb_003598_060 |
0.0243177483 |
- |
- |
hypothetical protein POPTR_0006s25370g [Populus trichocarpa] |
| 6 |
Hb_061183_010 |
0.0334760745 |
- |
- |
PREDICTED: uncharacterized protein LOC105775353 [Gossypium raimondii] |
| 7 |
Hb_001946_230 |
0.0409959336 |
- |
- |
- |
| 8 |
Hb_000313_160 |
0.0432091969 |
- |
- |
BnaC02g20570D [Brassica napus] |
| 9 |
Hb_000337_020 |
0.0495938657 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001281_030 |
0.0514697326 |
- |
- |
hypothetical protein POPTR_0010s18450g [Populus trichocarpa] |
| 11 |
Hb_000877_010 |
0.0515193685 |
- |
- |
PREDICTED: F-box protein At3g07870-like [Jatropha curcas] |
| 12 |
Hb_001068_060 |
0.0541548587 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 13 |
Hb_000032_120 |
0.0544236837 |
- |
- |
Bifunctional dihydroflavonol 4-reductase/flavanone 4-reductase [Glycine soja] |
| 14 |
Hb_099335_020 |
0.0560868351 |
- |
- |
hypothetical protein ARALYDRAFT_905869 [Arabidopsis lyrata subsp. lyrata] |
| 15 |
Hb_003685_020 |
0.0600009496 |
- |
- |
PREDICTED: laccase-14-like [Fragaria vesca subsp. vesca] |
| 16 |
Hb_034168_020 |
0.0629666746 |
- |
- |
putative uncharacterized protein (mitochondrion) [Hevea brasiliensis] |
| 17 |
Hb_118410_020 |
0.0679674214 |
- |
- |
PREDICTED: UDP-glycosyltransferase 76C4 [Vitis vinifera] |
| 18 |
Hb_001736_010 |
0.0822385117 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 19 |
Hb_124540_010 |
0.0894160963 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 3.1-like [Populus euphratica] |
| 20 |
Hb_010904_020 |
0.0917605137 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 [Populus euphratica] |