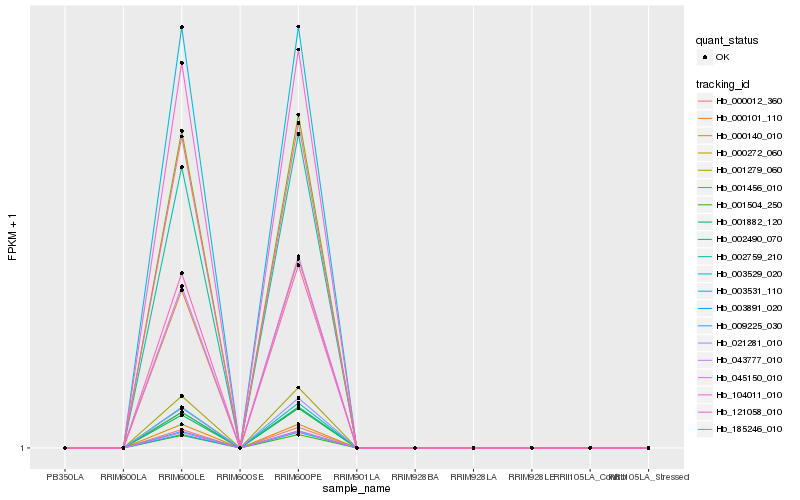
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002759_210 |
0.0 |
- |
- |
Peroxidase 6 precursor family protein [Populus trichocarpa] |
| 2 |
Hb_000012_360 |
0.001831797 |
- |
- |
PREDICTED: uncharacterized protein LOC104248159 [Nicotiana sylvestris] |
| 3 |
Hb_000272_060 |
0.0027540776 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_002490_070 |
0.0036468913 |
- |
- |
PREDICTED: uncharacterized protein LOC105123384 isoform X1 [Populus euphratica] |
| 5 |
Hb_003529_020 |
0.0048299057 |
- |
- |
hypothetical protein JCGZ_04979 [Jatropha curcas] |
| 6 |
Hb_001279_060 |
0.0064755749 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0001s36220g [Populus trichocarpa] |
| 7 |
Hb_003891_020 |
0.0069951331 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 2-1, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_045150_010 |
0.0076929711 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 9 |
Hb_043777_010 |
0.0121667418 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001504_250 |
0.0130866668 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_001456_010 |
0.0144829101 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000101_110 |
0.0146650419 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 13 |
Hb_121058_010 |
0.016146903 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_185246_010 |
0.0162023798 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 15 |
Hb_104011_010 |
0.0181018768 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Jatropha curcas] |
| 16 |
Hb_009225_030 |
0.0187238672 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 17 |
Hb_001882_120 |
0.0207318327 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 18 |
Hb_021281_010 |
0.0217725465 |
- |
- |
Major latex protein, putative [Ricinus communis] |
| 19 |
Hb_000140_010 |
0.0247757951 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 20 |
Hb_003531_110 |
0.0261147428 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |