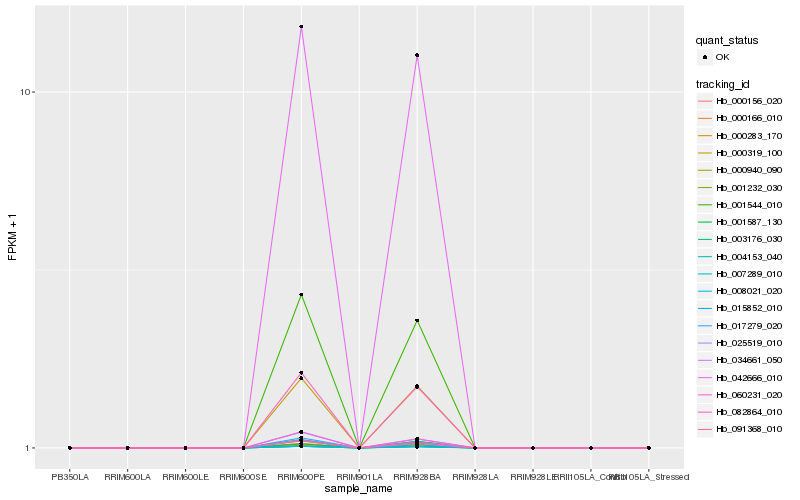
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001232_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s18270g [Populus trichocarpa] |
| 2 |
Hb_091368_010 |
0.0105503658 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 3 |
Hb_015852_010 |
0.0121359135 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 4 |
Hb_004153_040 |
0.0155869664 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_001544_010 |
0.0187090121 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000166_010 |
0.0236378142 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 7 |
Hb_082864_010 |
0.0238905508 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 8 |
Hb_017279_020 |
0.0255789329 |
- |
- |
protein MOTHER of FT and TF 1 [Jatropha curcas] |
| 9 |
Hb_042666_010 |
0.0357702995 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 10 |
Hb_034661_050 |
0.0383663355 |
- |
- |
PREDICTED: zinc finger MYM-type protein 1-like, partial [Malus domestica] |
| 11 |
Hb_000940_090 |
0.043671559 |
- |
- |
hypothetical protein CISIN_1g046520mg [Citrus sinensis] |
| 12 |
Hb_025519_010 |
0.0448058453 |
- |
- |
PREDICTED: uncharacterized protein LOC103485710 [Cucumis melo] |
| 13 |
Hb_000156_020 |
0.0455199507 |
- |
- |
PREDICTED: la-related protein 6A [Jatropha curcas] |
| 14 |
Hb_003176_030 |
0.0455199507 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Solanum lycopersicum] |
| 15 |
Hb_001587_130 |
0.0465871981 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 16 |
Hb_060231_020 |
0.0468379236 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22 [Jatropha curcas] |
| 17 |
Hb_000319_100 |
0.0469210826 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 18 |
Hb_000283_170 |
0.0553014837 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_008021_020 |
0.055614833 |
transcription factor |
TF Family: GRF |
hypothetical protein POPTR_0014s00780g [Populus trichocarpa] |
| 20 |
Hb_007289_010 |
0.0557668464 |
- |
- |
cytochrome P450, putative [Ricinus communis] |