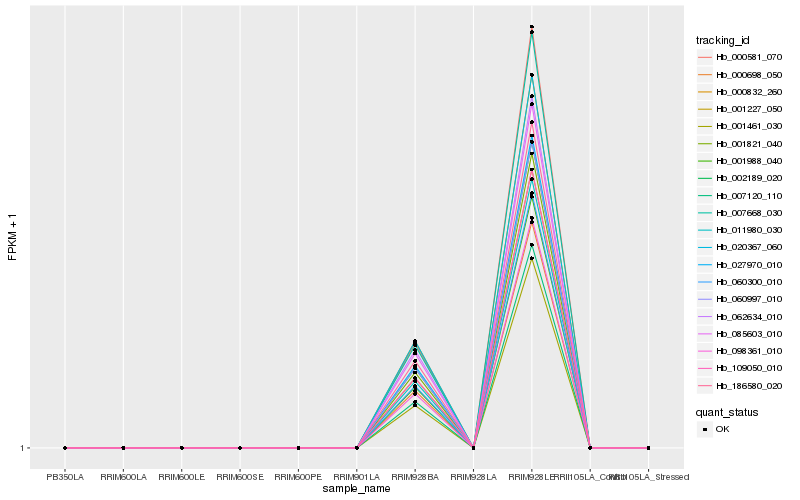
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000698_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_002189_020 |
0.0010126766 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_060997_010 |
0.0010126766 |
- |
- |
PREDICTED: uncharacterized protein LOC105789525 [Gossypium raimondii] |
| 4 |
Hb_001821_040 |
0.0011875037 |
- |
- |
- |
| 5 |
Hb_027970_010 |
0.0022013749 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 6 |
Hb_000832_260 |
0.0041048614 |
- |
- |
Protein gar2, putative [Ricinus communis] |
| 7 |
Hb_109050_010 |
0.0041048614 |
- |
- |
- |
| 8 |
Hb_186580_020 |
0.0047789671 |
- |
- |
PREDICTED: uncharacterized protein LOC103426965 [Malus domestica] |
| 9 |
Hb_011980_030 |
0.005834376 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_060300_010 |
0.0082769734 |
- |
- |
hypothetical protein F383_16817 [Gossypium arboreum] |
| 11 |
Hb_085603_010 |
0.0084409117 |
- |
- |
PREDICTED: uncharacterized protein LOC101296995 [Fragaria vesca subsp. vesca] |
| 12 |
Hb_001988_040 |
0.0088681556 |
- |
- |
hypothetical protein CICLE_v10006597mg [Citrus clementina] |
| 13 |
Hb_062634_010 |
0.0100661926 |
- |
- |
PREDICTED: uncharacterized protein LOC105636643 [Jatropha curcas] |
| 14 |
Hb_098361_010 |
0.012401556 |
- |
- |
PREDICTED: uncharacterized protein LOC105118398 [Populus euphratica] |
| 15 |
Hb_000581_070 |
0.0128602972 |
- |
- |
- |
| 16 |
Hb_007668_030 |
0.0133927724 |
- |
- |
PREDICTED: cold shock domain-containing protein 3-like [Musa acuminata subsp. malaccensis] |
| 17 |
Hb_001227_050 |
0.0146104043 |
- |
- |
- |
| 18 |
Hb_020367_060 |
0.0146104043 |
- |
- |
hypothetical protein POPTR_0005s15010g [Populus trichocarpa] |
| 19 |
Hb_007120_110 |
0.0166474562 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 20 |
Hb_001461_030 |
0.0187400544 |
- |
- |
- |