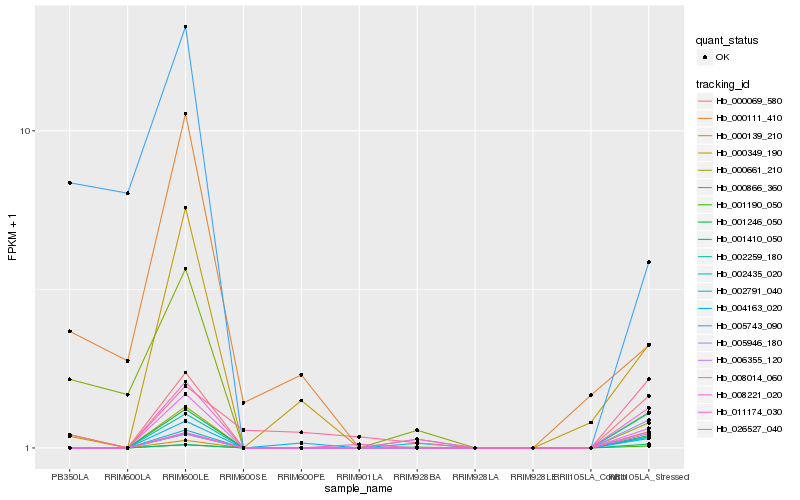
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000661_210 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_008221_020 |
0.2871318346 |
- |
- |
hypothetical protein JCGZ_15014 [Jatropha curcas] |
| 3 |
Hb_000069_580 |
0.287569077 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002435_020 |
0.2923244966 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 5 |
Hb_002259_180 |
0.3095038979 |
- |
- |
- |
| 6 |
Hb_001246_050 |
0.3125649137 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 7 |
Hb_006355_120 |
0.3164184575 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001410_050 |
0.3184672894 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 9 |
Hb_000349_190 |
0.3547709672 |
transcription factor |
TF Family: GRF |
hypothetical protein POPTR_0001s08310g [Populus trichocarpa] |
| 10 |
Hb_005743_090 |
0.3587318179 |
- |
- |
- |
| 11 |
Hb_000866_360 |
0.3651819602 |
- |
- |
- |
| 12 |
Hb_008014_060 |
0.3695009777 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 13 |
Hb_011174_030 |
0.3726965537 |
- |
- |
- |
| 14 |
Hb_001190_050 |
0.3772721805 |
- |
- |
hypothetical protein JCGZ_20709 [Jatropha curcas] |
| 15 |
Hb_004163_020 |
0.3797973864 |
- |
- |
- |
| 16 |
Hb_002791_040 |
0.380158975 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 17 |
Hb_026527_040 |
0.3842235067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000111_410 |
0.3863792957 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005946_180 |
0.3870261968 |
- |
- |
PREDICTED: uncharacterized protein LOC100818788 [Glycine max] |
| 20 |
Hb_000139_210 |
0.392468435 |
- |
- |
- |