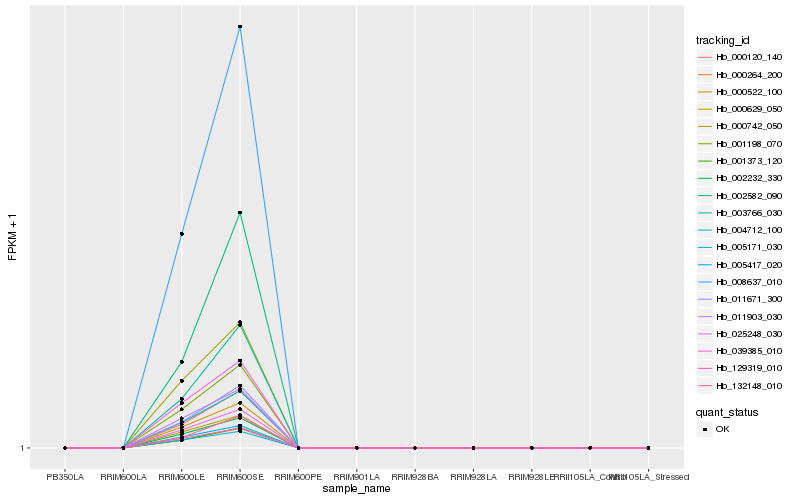
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000264_200 |
0.0 |
- |
- |
PREDICTED: tetraspanin-19-like [Jatropha curcas] |
| 2 |
Hb_000120_140 |
0.0002706976 |
- |
- |
hypothetical protein POPTR_0009s05160g [Populus trichocarpa] |
| 3 |
Hb_011671_300 |
0.0012207043 |
- |
- |
14.3 kDa oleosin [Jatropha curcas] |
| 4 |
Hb_008637_010 |
0.0064753144 |
- |
- |
Major latex protein, putative [Ricinus communis] |
| 5 |
Hb_001373_120 |
0.0107586238 |
- |
- |
Uncharacterized protein TCM_031358 [Theobroma cacao] |
| 6 |
Hb_004712_100 |
0.0150289945 |
- |
- |
hypothetical protein CICLE_v10003434mg, partial [Citrus clementina] |
| 7 |
Hb_003766_030 |
0.015157037 |
- |
- |
PREDICTED: nuclear transport factor 2-like [Gossypium raimondii] |
| 8 |
Hb_001198_070 |
0.020210866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000522_100 |
0.0246181422 |
- |
- |
PREDICTED: putative SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3-like 3 [Nelumbo nucifera] |
| 10 |
Hb_002232_330 |
0.0248268426 |
- |
- |
- |
| 11 |
Hb_025248_030 |
0.026492467 |
- |
- |
hypothetical protein JCGZ_09377 [Jatropha curcas] |
| 12 |
Hb_005171_030 |
0.0361840203 |
- |
- |
hypothetical protein EUGRSUZ_H02434 [Eucalyptus grandis] |
| 13 |
Hb_129319_010 |
0.036389889 |
- |
- |
PREDICTED: uncharacterized protein LOC104115096 [Nicotiana tomentosiformis] |
| 14 |
Hb_005417_020 |
0.0365784373 |
- |
- |
- |
| 15 |
Hb_000629_050 |
0.0372982646 |
- |
- |
- |
| 16 |
Hb_011903_030 |
0.0392577575 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 [Populus euphratica] |
| 17 |
Hb_132148_010 |
0.0406807063 |
- |
- |
hypothetical protein CICLE_v10006890mg, partial [Citrus clementina] |
| 18 |
Hb_039385_010 |
0.0415126521 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_000742_050 |
0.0448518559 |
- |
- |
hypothetical protein CL1373Contig1_03, partial [Pinus lambertiana] |
| 20 |
Hb_002582_090 |
0.0503625811 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like [Sesamum indicum] |