| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
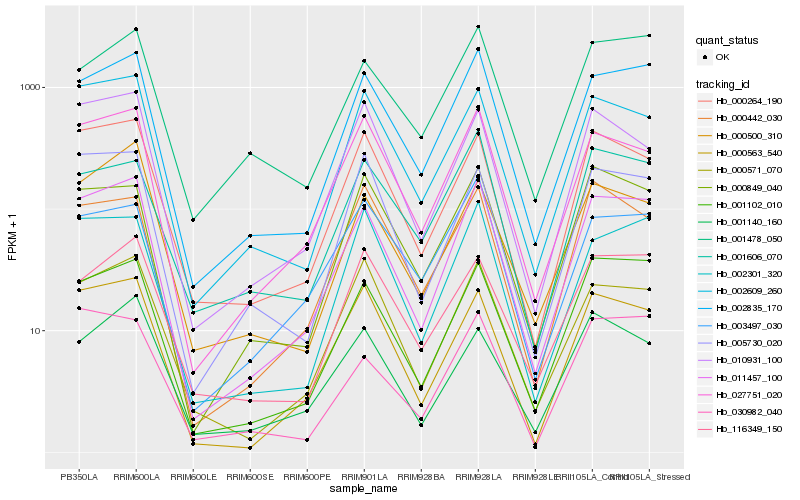
Hb_011457_100 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_002835_170 |
0.0636628772 |
- |
- |
PREDICTED: uncharacterized protein LOC105640307 [Jatropha curcas] |
| 3 |
Hb_001102_010 |
0.0833376281 |
- |
- |
PREDICTED: solanesyl diphosphate synthase 3, chloroplastic/mitochondrial-like isoform X2 [Glycine max] |
| 4 |
Hb_027751_020 |
0.0957520012 |
- |
- |
PREDICTED: uncharacterized protein LOC104886652 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_000563_540 |
0.0973998811 |
- |
- |
PREDICTED: Fanconi anemia group I protein [Jatropha curcas] |
| 6 |
Hb_002609_260 |
0.100145631 |
- |
- |
CCCH-type zinc finger protein [Hevea brasiliensis] |
| 7 |
Hb_000264_190 |
0.1029628099 |
- |
- |
sphingolipid delta 4 desaturase/C-4 hydroxylase protein des2, putative [Ricinus communis] |
| 8 |
Hb_000571_070 |
0.1071597577 |
transcription factor |
TF Family: G2-like |
PREDICTED: putative Myb family transcription factor At1g14600 [Jatropha curcas] |
| 9 |
Hb_002301_320 |
0.1130363728 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0004s04590g [Populus trichocarpa] |
| 10 |
Hb_001478_050 |
0.1175322281 |
- |
- |
PREDICTED: uncharacterized protein LOC105640307 [Jatropha curcas] |
| 11 |
Hb_005730_020 |
0.1192605413 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.1-like [Jatropha curcas] |
| 12 |
Hb_000849_040 |
0.1209825981 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_010931_100 |
0.1248178701 |
- |
- |
PREDICTED: uncharacterized protein LOC105638669 [Jatropha curcas] |
| 14 |
Hb_116349_150 |
0.1258701952 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 15 |
Hb_030982_040 |
0.1282096286 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 16 |
Hb_001606_070 |
0.1289287444 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 17 |
Hb_000500_310 |
0.129476284 |
- |
- |
- |
| 18 |
Hb_003497_030 |
0.1301091132 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 19 |
Hb_001140_160 |
0.1318614559 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 20 |
Hb_000442_030 |
0.1327041246 |
- |
- |
conserved hypothetical protein [Ricinus communis] |