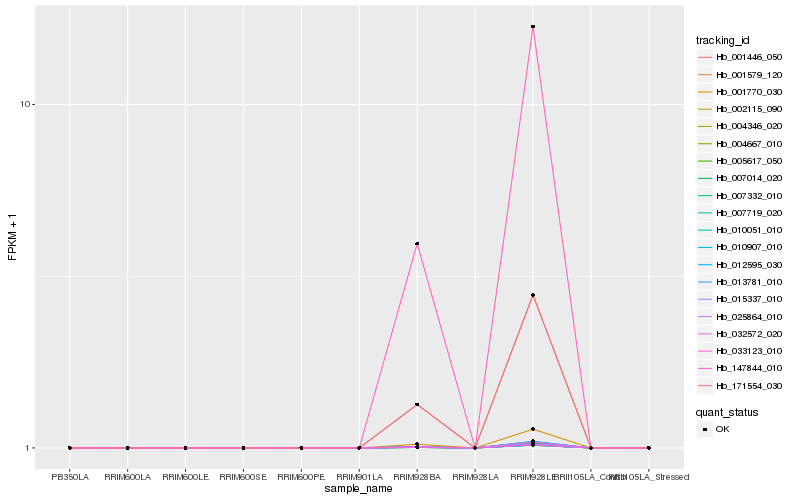
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005617_050 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105167812 [Sesamum indicum] |
| 2 |
Hb_001579_120 |
0.000166689 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_001770_030 |
0.000263637 |
- |
- |
hypothetical protein B456_010G041100, partial [Gossypium raimondii] |
| 4 |
Hb_015337_010 |
0.0005325655 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_013781_010 |
0.0005674371 |
- |
- |
hypothetical protein VITISV_030726 [Vitis vinifera] |
| 6 |
Hb_007332_010 |
0.0010098117 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 7 |
Hb_033123_010 |
0.0012903687 |
- |
- |
Putative polyprotein [Oryza sativa Japonica Group] |
| 8 |
Hb_010051_010 |
0.001339338 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 9 |
Hb_001446_050 |
0.0013649757 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 10 |
Hb_002115_090 |
0.0013834302 |
- |
- |
PREDICTED: uncharacterized protein LOC104590556 [Nelumbo nucifera] |
| 11 |
Hb_032572_020 |
0.0014271296 |
- |
- |
- |
| 12 |
Hb_004346_020 |
0.0015701857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_171554_030 |
0.0015701857 |
- |
- |
- |
| 14 |
Hb_025864_010 |
0.0016481891 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 15 |
Hb_007014_020 |
0.0017497332 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 16 |
Hb_147844_010 |
0.0017659371 |
- |
- |
putative 60S ribosomal protein L37 [Aegilops tauschii] |
| 17 |
Hb_010907_010 |
0.0018603722 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 18 |
Hb_012595_030 |
0.001906286 |
- |
- |
- |
| 19 |
Hb_007719_020 |
0.001935672 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 20 |
Hb_004667_010 |
0.0019630712 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |