| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
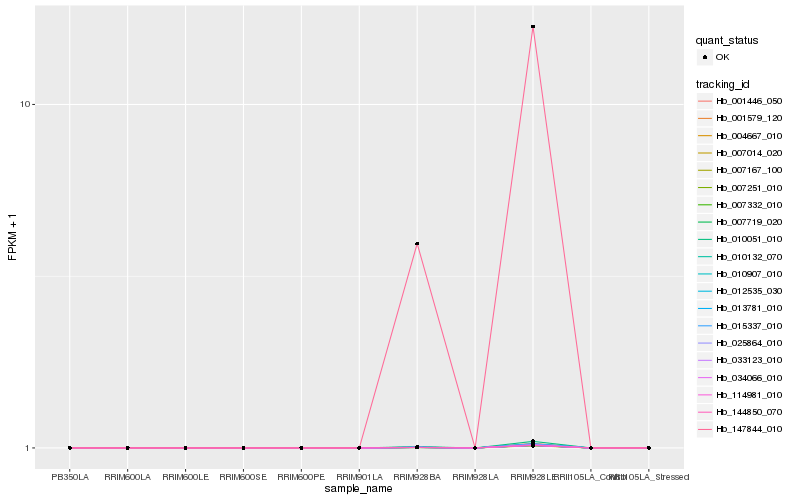
Hb_033123_010 |
0.0 |
- |
- |
Putative polyprotein [Oryza sativa Japonica Group] |
| 2 |
Hb_010051_010 |
4.89694e-05 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 3 |
Hb_001446_050 |
7.46072e-05 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 4 |
Hb_007332_010 |
0.0002805573 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 5 |
Hb_025864_010 |
0.0003578212 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_007014_020 |
0.0004593655 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 7 |
Hb_147844_010 |
0.0004755695 |
- |
- |
putative 60S ribosomal protein L37 [Aegilops tauschii] |
| 8 |
Hb_010907_010 |
0.0005700048 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 9 |
Hb_007719_020 |
0.0006453048 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 10 |
Hb_004667_010 |
0.0006727042 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_114981_010 |
0.0007120292 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 12 |
Hb_013781_010 |
0.0007229321 |
- |
- |
hypothetical protein VITISV_030726 [Vitis vinifera] |
| 13 |
Hb_015337_010 |
0.0007578037 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_012535_030 |
0.0007780338 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 15 |
Hb_144850_070 |
0.0007862303 |
- |
- |
OSJNBa0032N05.11 [Oryza sativa Japonica Group] |
| 16 |
Hb_034066_010 |
0.0008312258 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_010132_070 |
0.000859194 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_007251_010 |
0.0009791461 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1-like [Jatropha curcas] |
| 19 |
Hb_001579_120 |
0.0011236799 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_007167_100 |
0.0012070553 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |