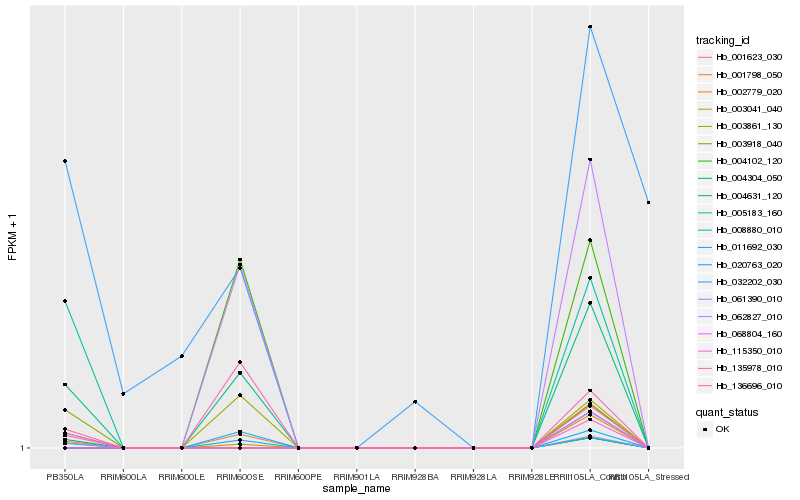
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004631_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_011692_030 |
0.3400902564 |
- |
- |
PREDICTED: uncharacterized protein LOC105775392, partial [Gossypium raimondii] |
| 3 |
Hb_020763_020 |
0.3401375122 |
- |
- |
hypothetical protein POPTR_0001s27010g [Populus trichocarpa] |
| 4 |
Hb_062827_010 |
0.3410373979 |
- |
- |
- |
| 5 |
Hb_003041_040 |
0.3434621477 |
- |
- |
PREDICTED: uncharacterized protein LOC104908276 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_004102_120 |
0.3599250737 |
- |
- |
- |
| 7 |
Hb_003861_130 |
0.3757952763 |
- |
- |
- |
| 8 |
Hb_068804_160 |
0.3758213115 |
transcription factor |
TF Family: MIKC |
hypothetical protein JCGZ_12632 [Jatropha curcas] |
| 9 |
Hb_136696_010 |
0.376101278 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 10 |
Hb_002779_020 |
0.3761325846 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_115350_010 |
0.3761500944 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 12 |
Hb_061390_010 |
0.3762134372 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 13 |
Hb_008880_010 |
0.3762190043 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 isoform X2 [Nelumbo nucifera] |
| 14 |
Hb_001623_030 |
0.3919167881 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 15 |
Hb_004304_050 |
0.3994513751 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 16 |
Hb_005183_160 |
0.4008621103 |
- |
- |
- |
| 17 |
Hb_003918_040 |
0.4029284728 |
- |
- |
- |
| 18 |
Hb_001798_050 |
0.4066831543 |
transcription factor |
TF Family: NAC |
NAC transcription factor 057 [Jatropha curcas] |
| 19 |
Hb_135978_010 |
0.4085361841 |
- |
- |
hypothetical protein EUGRSUZ_J026621, partial [Eucalyptus grandis] |
| 20 |
Hb_032202_030 |
0.4158200364 |
- |
- |
- |