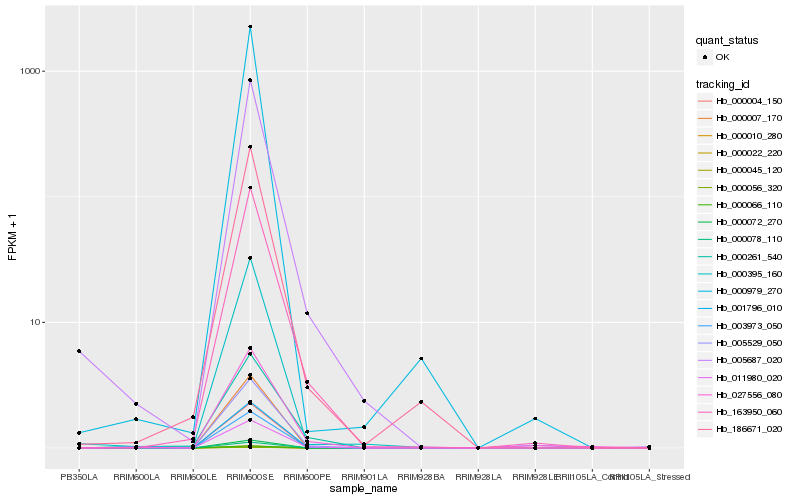
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003973_050 |
0.0 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 3-like isoform X1 [Populus euphratica] |
| 2 |
Hb_005687_020 |
0.0990344022 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 3 |
Hb_011980_020 |
0.1034049392 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_163950_060 |
0.1062694953 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 5 |
Hb_000395_160 |
0.1110506159 |
- |
- |
hypothetical protein POPTR_0006s25490g [Populus trichocarpa] |
| 6 |
Hb_186671_020 |
0.1111446303 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_027556_080 |
0.1129046036 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g24130 [Jatropha curcas] |
| 8 |
Hb_001796_010 |
0.11668811 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 9 |
Hb_000261_540 |
0.1171672595 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1-like [Jatropha curcas] |
| 10 |
Hb_005529_050 |
0.1196762247 |
- |
- |
PREDICTED: cytochrome P450 94B3-like [Nelumbo nucifera] |
| 11 |
Hb_000979_270 |
0.1198542914 |
- |
- |
PREDICTED: MLP-like protein 31 [Jatropha curcas] |
| 12 |
Hb_000004_150 |
0.1206440719 |
- |
- |
hypothetical protein RCOM_1382630 [Ricinus communis] |
| 13 |
Hb_000007_170 |
0.1206440719 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 2-like protein [Nelumbo nucifera] |
| 14 |
Hb_000010_280 |
0.1206440719 |
- |
- |
hypothetical protein JCGZ_13805 [Jatropha curcas] |
| 15 |
Hb_000022_220 |
0.1206440719 |
- |
- |
PREDICTED: potassium channel KAT3 [Jatropha curcas] |
| 16 |
Hb_000045_120 |
0.1206440719 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 17 |
Hb_000056_320 |
0.1206440719 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000066_110 |
0.1206440719 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01370 [Jatropha curcas] |
| 19 |
Hb_000072_270 |
0.1206440719 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 20 |
Hb_000078_110 |
0.1206440719 |
- |
- |
PREDICTED: uncharacterized protein LOC105640496 [Jatropha curcas] |