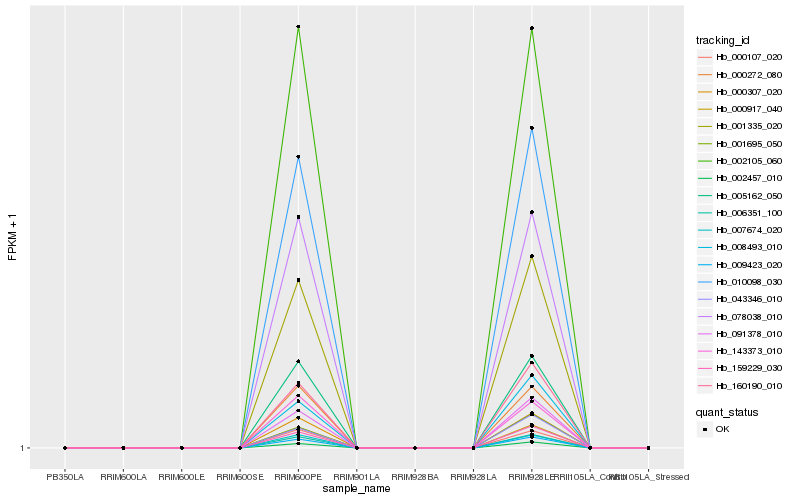
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000307_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC100255757 [Vitis vinifera] |
| 2 |
Hb_010098_030 |
0.0028275237 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000917_040 |
0.0070421648 |
- |
- |
PREDICTED: uncharacterized protein LOC105639996 [Jatropha curcas] |
| 4 |
Hb_001335_020 |
0.0073422526 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 5 |
Hb_006351_100 |
0.0097699271 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 6 |
Hb_005162_050 |
0.0159737522 |
- |
- |
hypothetical protein B456_009G352100 [Gossypium raimondii] |
| 7 |
Hb_009423_020 |
0.0162205087 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_078038_010 |
0.0247330848 |
- |
- |
hypothetical protein CISIN_1g038197mg [Citrus sinensis] |
| 9 |
Hb_002105_060 |
0.0331811107 |
- |
- |
hypothetical protein POPTR_0022s00320g [Populus trichocarpa] |
| 10 |
Hb_000272_080 |
0.0345877264 |
- |
- |
temperature-induced lipocalin [Populus tremula x Populus tremuloides] |
| 11 |
Hb_160190_010 |
0.0347791063 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 12 |
Hb_002457_010 |
0.0359118478 |
- |
- |
polyprotein [Oryza australiensis] |
| 13 |
Hb_091378_010 |
0.0371866468 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 14 |
Hb_007674_020 |
0.0374940218 |
- |
- |
PREDICTED: serpin-ZX-like [Jatropha curcas] |
| 15 |
Hb_159229_030 |
0.037844325 |
- |
- |
hypothetical protein CISIN_1g0034891mg, partial [Citrus sinensis] |
| 16 |
Hb_000107_020 |
0.0458856343 |
- |
- |
PREDICTED: uncharacterized protein LOC105032542 [Elaeis guineensis] |
| 17 |
Hb_143373_010 |
0.0595415617 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 [Gossypium raimondii] |
| 18 |
Hb_008493_010 |
0.0725527747 |
- |
- |
hypothetical protein JCGZ_10529 [Jatropha curcas] |
| 19 |
Hb_001695_050 |
0.0796143169 |
- |
- |
PREDICTED: uncharacterized protein LOC104884077 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_043346_010 |
0.0825318324 |
- |
- |
PREDICTED: uncharacterized protein LOC105761922 [Gossypium raimondii] |