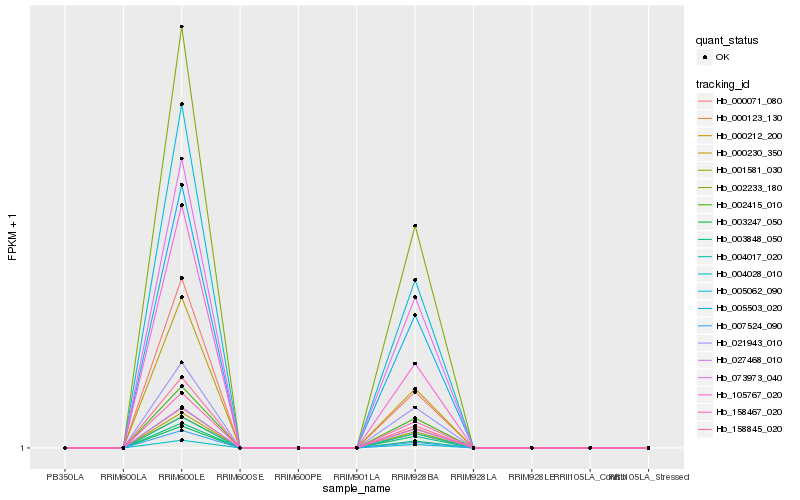
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000230_350 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630371 [Jatropha curcas] |
| 2 |
Hb_105767_020 |
0.0289891078 |
- |
- |
- |
| 3 |
Hb_007524_090 |
0.0312322493 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 4 |
Hb_005062_090 |
0.0337730984 |
- |
- |
ferredoxin [Vibrio natriegens] |
| 5 |
Hb_003247_050 |
0.0342338705 |
- |
- |
- |
| 6 |
Hb_000071_080 |
0.0347157648 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO5 [Jatropha curcas] |
| 7 |
Hb_004028_010 |
0.0355771445 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 8 |
Hb_000212_200 |
0.0368090764 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105645015 [Jatropha curcas] |
| 9 |
Hb_001581_030 |
0.037020882 |
- |
- |
PREDICTED: putative F-box/FBD/LRR-repeat protein At5g56810 [Jatropha curcas] |
| 10 |
Hb_027468_010 |
0.0373098798 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_158845_020 |
0.0388820102 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like protein 8 [Jatropha curcas] |
| 12 |
Hb_021943_010 |
0.0397739718 |
- |
- |
- |
| 13 |
Hb_004017_020 |
0.0411032679 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 14 |
Hb_003848_050 |
0.0417498504 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |
| 15 |
Hb_158467_020 |
0.0439134159 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 16 |
Hb_002415_010 |
0.0443915534 |
- |
- |
PREDICTED: uncharacterized protein LOC105775231 [Gossypium raimondii] |
| 17 |
Hb_005503_020 |
0.0446818236 |
- |
- |
Hypothetical protein glysoja_025074 [Glycine soja] |
| 18 |
Hb_002233_180 |
0.0464585765 |
- |
- |
- |
| 19 |
Hb_000123_130 |
0.0485958312 |
- |
- |
hypothetical protein POPTR_0008s05200g, partial [Populus trichocarpa] |
| 20 |
Hb_073973_040 |
0.049898691 |
- |
- |
PREDICTED: uncharacterized protein LOC103941330 [Pyrus x bretschneideri] |