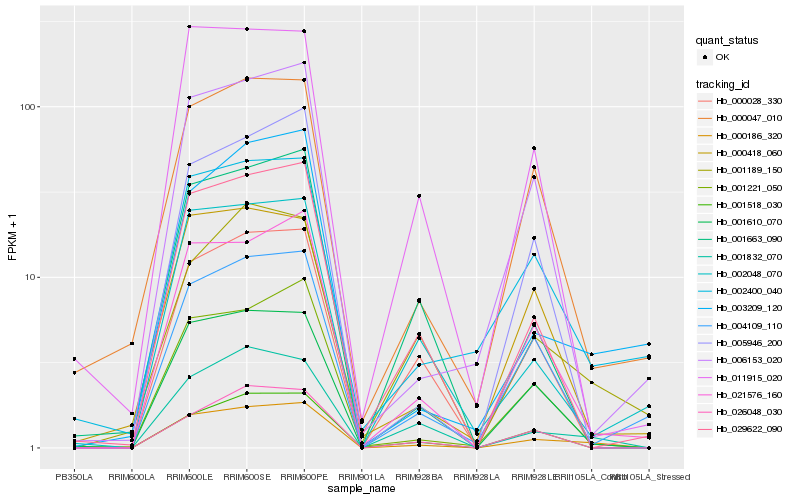
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000186_320 |
0.0 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 2 |
Hb_001832_070 |
0.1212636993 |
- |
- |
PREDICTED: uncharacterized protein LOC105637887 [Jatropha curcas] |
| 3 |
Hb_021576_160 |
0.1212969701 |
transcription factor |
TF Family: WRKY |
putative WRKY transcription factor family protein [Populus trichocarpa] |
| 4 |
Hb_001221_050 |
0.1357618656 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |
| 5 |
Hb_001610_070 |
0.1375921958 |
- |
- |
hypothetical protein MIMGU_mgv1a022867mg [Erythranthe guttata] |
| 6 |
Hb_029622_090 |
0.1398916246 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 2-like [Jatropha curcas] |
| 7 |
Hb_000028_330 |
0.1485805894 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 9-like [Pyrus x bretschneideri] |
| 8 |
Hb_000047_010 |
0.1541116966 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 9 |
Hb_001189_150 |
0.1543312737 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 89-like [Jatropha curcas] |
| 10 |
Hb_005946_200 |
0.1549075213 |
- |
- |
hypothetical protein POPTR_0006s16210g [Populus trichocarpa] |
| 11 |
Hb_006153_020 |
0.1552910624 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 12 |
Hb_001663_090 |
0.1556389059 |
- |
- |
ribonuclease t2, putative [Ricinus communis] |
| 13 |
Hb_001518_030 |
0.1569696309 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 14 |
Hb_004109_110 |
0.1570915613 |
- |
- |
dioxygenase family protein [Populus trichocarpa] |
| 15 |
Hb_011915_020 |
0.1578122893 |
- |
- |
PREDICTED: arogenate dehydratase/prephenate dehydratase 6, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_003209_120 |
0.1587104131 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 17 |
Hb_002048_070 |
0.1588846457 |
- |
- |
hypothetical protein JCGZ_15146 [Jatropha curcas] |
| 18 |
Hb_002400_040 |
0.1599760344 |
- |
- |
PREDICTED: uncharacterized protein LOC105637633 [Jatropha curcas] |
| 19 |
Hb_026048_030 |
0.1614807921 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000418_060 |
0.1663082993 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |