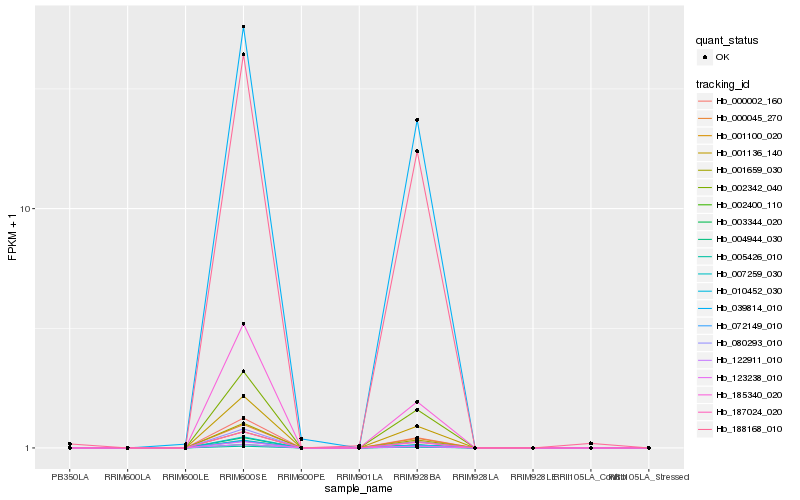
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_187024_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC102598429 [Solanum tuberosum] |
| 2 |
Hb_000002_160 |
0.0086696314 |
- |
- |
Uncharacterized protein TCM_021030 [Theobroma cacao] |
| 3 |
Hb_080293_010 |
0.0130115461 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Nelumbo nucifera] |
| 4 |
Hb_001136_140 |
0.0172154372 |
- |
- |
hypothetical protein POPTR_0010s13790g [Populus trichocarpa] |
| 5 |
Hb_001659_030 |
0.0177544869 |
- |
- |
- |
| 6 |
Hb_004944_030 |
0.0180681698 |
- |
- |
PREDICTED: uncharacterized protein LOC104227769, partial [Nicotiana sylvestris] |
| 7 |
Hb_003344_020 |
0.0192078451 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103501197 [Cucumis melo] |
| 8 |
Hb_072149_010 |
0.0197661689 |
- |
- |
hypothetical protein CICLE_v10006890mg, partial [Citrus clementina] |
| 9 |
Hb_002400_110 |
0.0200082467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_123238_010 |
0.0212303812 |
- |
- |
hypothetical protein JCGZ_16598 [Jatropha curcas] |
| 11 |
Hb_001100_020 |
0.0251881061 |
- |
- |
- |
| 12 |
Hb_010452_030 |
0.0265056209 |
transcription factor |
TF Family: MYB |
Myb domain protein 107 [Theobroma cacao] |
| 13 |
Hb_000045_270 |
0.0291968309 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_122911_010 |
0.0348806012 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial [Jatropha curcas] |
| 15 |
Hb_188168_010 |
0.0407128328 |
- |
- |
Arylacetamide deacetylase, putative [Ricinus communis] |
| 16 |
Hb_002342_040 |
0.0420786584 |
- |
- |
FERONIA, partial [Arabis alpina] |
| 17 |
Hb_039814_010 |
0.0462379248 |
- |
- |
hypothetical protein JCGZ_05023 [Jatropha curcas] |
| 18 |
Hb_007259_030 |
0.0520882421 |
- |
- |
- |
| 19 |
Hb_185340_020 |
0.0523707242 |
- |
- |
- |
| 20 |
Hb_005426_010 |
0.0524532817 |
- |
- |
polyprotein [Oryza australiensis] |