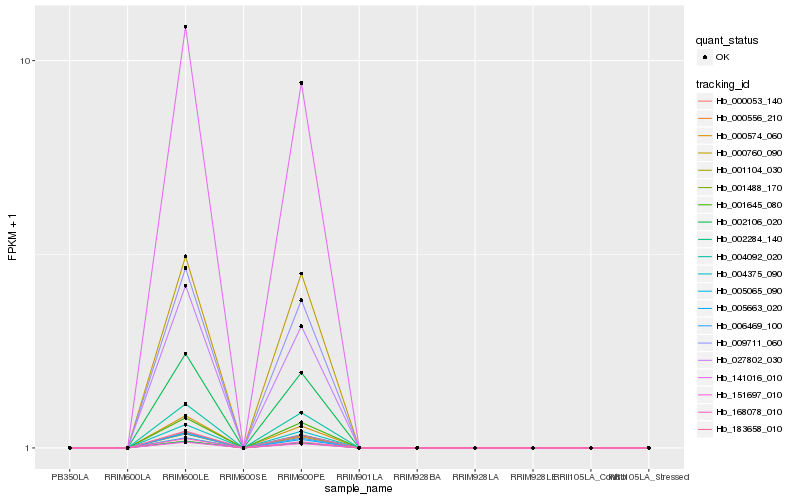
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_183658_010 |
0.0 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 2 |
Hb_151697_010 |
0.0011044037 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 3 |
Hb_009711_060 |
0.0013659228 |
- |
- |
- |
| 4 |
Hb_002106_020 |
0.0029543331 |
- |
- |
PREDICTED: gibberellic acid methyltransferase 2 [Vitis vinifera] |
| 5 |
Hb_000556_210 |
0.0055405824 |
- |
- |
hypothetical protein POPTR_0014s06640g [Populus trichocarpa] |
| 6 |
Hb_001104_030 |
0.0083097689 |
- |
- |
PREDICTED: uncharacterized protein LOC105125581 [Populus euphratica] |
| 7 |
Hb_002284_140 |
0.010639396 |
- |
- |
hypothetical protein POPTR_0002s00800g [Populus trichocarpa] |
| 8 |
Hb_004092_020 |
0.0110364836 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 32 [Jatropha curcas] |
| 9 |
Hb_004375_090 |
0.0115727936 |
- |
- |
hypothetical protein JCGZ_13971 [Jatropha curcas] |
| 10 |
Hb_141016_010 |
0.0149973039 |
- |
- |
hypothetical protein JCGZ_15085 [Jatropha curcas] |
| 11 |
Hb_000053_140 |
0.0206119675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_006469_100 |
0.0234822051 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 13 |
Hb_005663_020 |
0.0246876331 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 14 |
Hb_000574_060 |
0.0250271441 |
- |
- |
hypothetical protein F775_19731 [Aegilops tauschii] |
| 15 |
Hb_001645_080 |
0.0252160423 |
transcription factor |
TF Family: C2H2 |
palmate-like pentafoliata 1 transcription factor [Manihot esculenta] |
| 16 |
Hb_027802_030 |
0.0260899502 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0017s11800g [Populus trichocarpa] |
| 17 |
Hb_001488_170 |
0.0272138308 |
- |
- |
PREDICTED: transcription initiation factor IIB-like [Jatropha curcas] |
| 18 |
Hb_168078_010 |
0.0287818643 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like, partial [Jatropha curcas] |
| 19 |
Hb_000760_090 |
0.0306654452 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-6 [Vitis vinifera] |
| 20 |
Hb_005065_090 |
0.0309066476 |
- |
- |
zinc/iron transporter, putative [Ricinus communis] |