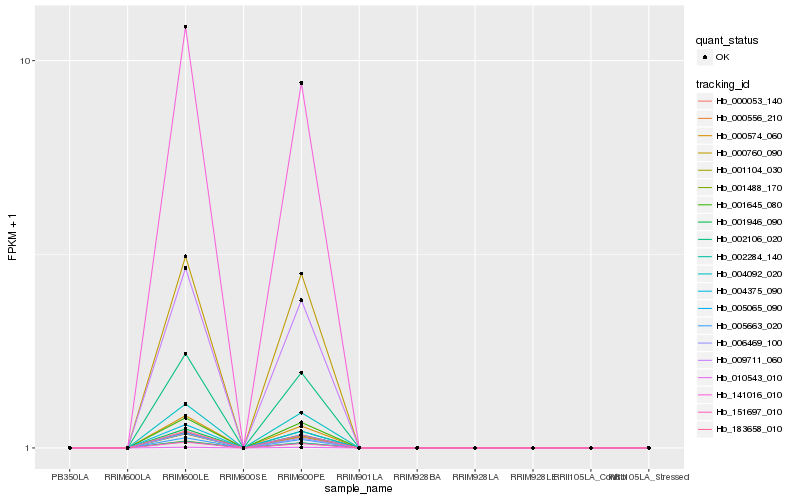
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000556_210 |
0.0 |
- |
- |
hypothetical protein POPTR_0014s06640g [Populus trichocarpa] |
| 2 |
Hb_002106_020 |
0.0025862646 |
- |
- |
PREDICTED: gibberellic acid methyltransferase 2 [Vitis vinifera] |
| 3 |
Hb_002284_140 |
0.0050989217 |
- |
- |
hypothetical protein POPTR_0002s00800g [Populus trichocarpa] |
| 4 |
Hb_004092_020 |
0.005496022 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 32 [Jatropha curcas] |
| 5 |
Hb_183658_010 |
0.0055405824 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 6 |
Hb_151697_010 |
0.0066449714 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 7 |
Hb_009711_060 |
0.0069064863 |
- |
- |
- |
| 8 |
Hb_001104_030 |
0.01385012 |
- |
- |
PREDICTED: uncharacterized protein LOC105125581 [Populus euphratica] |
| 9 |
Hb_000053_140 |
0.0150720016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004375_090 |
0.0171129774 |
- |
- |
hypothetical protein JCGZ_13971 [Jatropha curcas] |
| 11 |
Hb_005663_020 |
0.0191479873 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 12 |
Hb_001645_080 |
0.0196764428 |
transcription factor |
TF Family: C2H2 |
palmate-like pentafoliata 1 transcription factor [Manihot esculenta] |
| 13 |
Hb_141016_010 |
0.0205372654 |
- |
- |
hypothetical protein JCGZ_15085 [Jatropha curcas] |
| 14 |
Hb_001488_170 |
0.021674416 |
- |
- |
PREDICTED: transcription initiation factor IIB-like [Jatropha curcas] |
| 15 |
Hb_000760_090 |
0.025126386 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-6 [Vitis vinifera] |
| 16 |
Hb_005065_090 |
0.0253676149 |
- |
- |
zinc/iron transporter, putative [Ricinus communis] |
| 17 |
Hb_006469_100 |
0.0290214078 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 18 |
Hb_001946_090 |
0.0304318848 |
- |
- |
- |
| 19 |
Hb_000574_060 |
0.0305661766 |
- |
- |
hypothetical protein F775_19731 [Aegilops tauschii] |
| 20 |
Hb_010543_010 |
0.0314744957 |
- |
- |
Phospholipid-transporting ATPase, putative [Ricinus communis] |