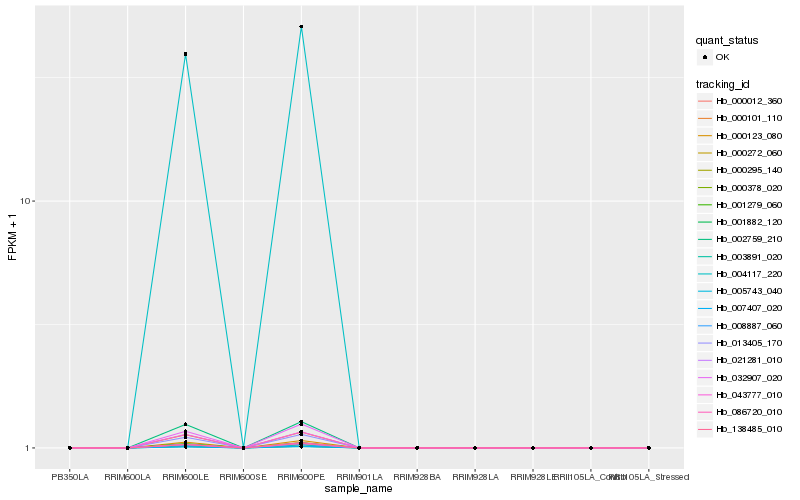
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013405_170 |
0.0 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000123_080 |
0.0006495565 |
transcription factor |
TF Family: B3 |
hypothetical protein VITISV_040593 [Vitis vinifera] |
| 3 |
Hb_005743_040 |
0.0008622013 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 4 |
Hb_000378_020 |
0.001719796 |
- |
- |
PREDICTED: uncharacterized protein LOC101497268 [Cicer arietinum] |
| 5 |
Hb_004117_220 |
0.0019623551 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 1-like [Jatropha curcas] |
| 6 |
Hb_021281_010 |
0.005497678 |
- |
- |
Major latex protein, putative [Ricinus communis] |
| 7 |
Hb_001882_120 |
0.0065385444 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 8 |
Hb_008887_060 |
0.0079078198 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000101_110 |
0.0126058072 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 10 |
Hb_043777_010 |
0.0151040955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_138485_010 |
0.0173463964 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 12 |
Hb_003891_020 |
0.0202752989 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 2-1, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_001279_060 |
0.0207947881 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0001s36220g [Populus trichocarpa] |
| 14 |
Hb_007407_020 |
0.0251531196 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_086720_010 |
0.0262119136 |
- |
- |
CTV.19 [Citrus trifoliata] |
| 16 |
Hb_002759_210 |
0.0272690702 |
- |
- |
Peroxidase 6 precursor family protein [Populus trichocarpa] |
| 17 |
Hb_000295_140 |
0.028751733 |
- |
- |
PREDICTED: uncharacterized protein LOC105628741 [Jatropha curcas] |
| 18 |
Hb_000012_360 |
0.0291003565 |
- |
- |
PREDICTED: uncharacterized protein LOC104248159 [Nicotiana sylvestris] |
| 19 |
Hb_032907_020 |
0.0295806292 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 20 |
Hb_000272_060 |
0.0300223559 |
- |
- |
unnamed protein product [Vitis vinifera] |