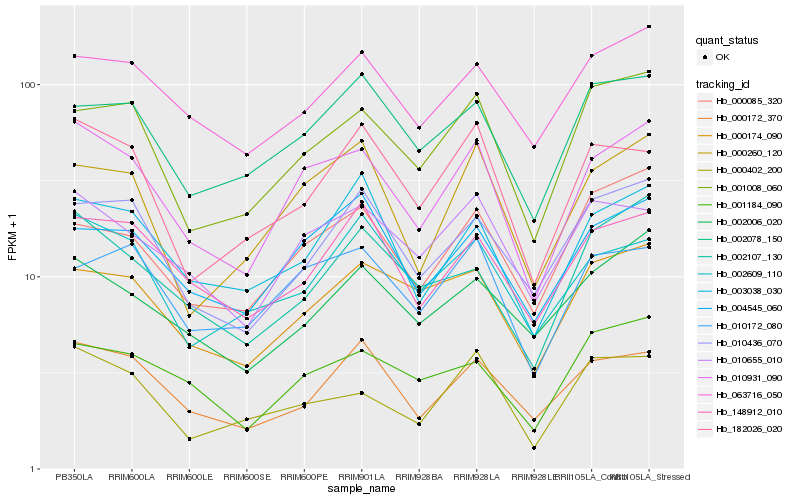
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010931_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_010655_010 |
0.1005140848 |
- |
- |
Cycloartenol synthase, putative [Ricinus communis] |
| 3 |
Hb_000260_120 |
0.1034616199 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_001008_060 |
0.1038178733 |
- |
- |
PREDICTED: monothiol glutaredoxin-S15, mitochondrial [Jatropha curcas] |
| 5 |
Hb_004545_060 |
0.1039368394 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 6B [Jatropha curcas] |
| 6 |
Hb_003038_030 |
0.1110355913 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 10 [Cucumis melo] |
| 7 |
Hb_010172_080 |
0.1110702203 |
- |
- |
acetylglucosaminyltransferase, putative [Ricinus communis] |
| 8 |
Hb_063716_050 |
0.1115075344 |
- |
- |
PREDICTED: uncharacterized protein LOC105634866 [Jatropha curcas] |
| 9 |
Hb_000174_090 |
0.1116160745 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 10 |
Hb_000172_370 |
0.1122063801 |
- |
- |
- |
| 11 |
Hb_000402_200 |
0.1131124938 |
- |
- |
PREDICTED: uncharacterized protein LOC105644655 [Jatropha curcas] |
| 12 |
Hb_001184_090 |
0.1135950425 |
- |
- |
PREDICTED: TIP41-like protein [Jatropha curcas] |
| 13 |
Hb_000085_320 |
0.113821973 |
- |
- |
Uncharacterized protein TCM_030749 [Theobroma cacao] |
| 14 |
Hb_148912_010 |
0.1144225405 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_010436_070 |
0.1149354715 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 4-like [Nicotiana tomentosiformis] |
| 16 |
Hb_182026_020 |
0.1158972492 |
- |
- |
PREDICTED: SUN domain-containing protein 3-like [Jatropha curcas] |
| 17 |
Hb_002107_130 |
0.1164276295 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_002006_020 |
0.1172023122 |
- |
- |
PREDICTED: uncharacterized protein LOC105650638 [Jatropha curcas] |
| 19 |
Hb_002078_150 |
0.1177778194 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_002609_110 |
0.1192577992 |
- |
- |
hypothetical protein JCGZ_08497 [Jatropha curcas] |