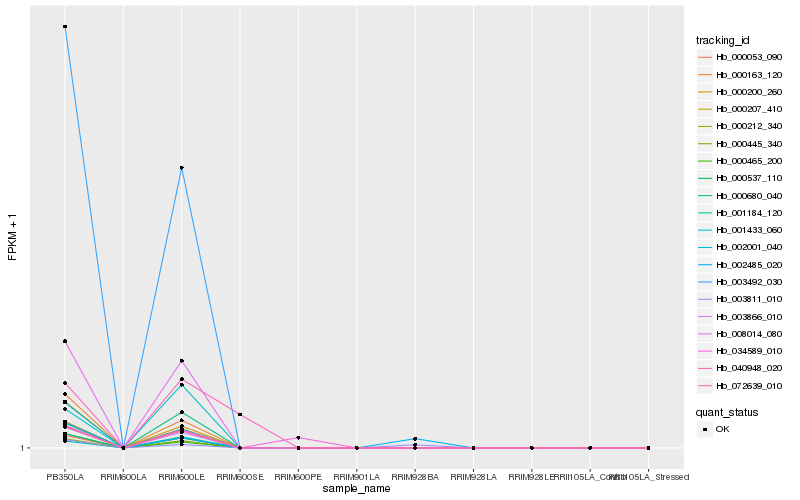
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003492_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_000163_120 |
0.0306840295 |
- |
- |
- |
| 3 |
Hb_003866_010 |
0.0532695446 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 4 |
Hb_000445_340 |
0.0620555366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000465_200 |
0.0657725021 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 1 [Jatropha curcas] |
| 6 |
Hb_000053_090 |
0.065905305 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Populus euphratica] |
| 7 |
Hb_072639_010 |
0.0660778281 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 8 |
Hb_008014_080 |
0.0671759065 |
- |
- |
PREDICTED: uncharacterized protein LOC103848614 [Brassica rapa] |
| 9 |
Hb_000212_340 |
0.0682699278 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000200_260 |
0.0721563265 |
- |
- |
PREDICTED: uncharacterized protein LOC104232531 isoform X2 [Nicotiana sylvestris] |
| 11 |
Hb_002001_040 |
0.0722733724 |
- |
- |
hypothetical protein CISIN_1g044945mg, partial [Citrus sinensis] |
| 12 |
Hb_000207_410 |
0.0740179863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000537_110 |
0.074779725 |
- |
- |
- |
| 14 |
Hb_001184_120 |
0.2150945916 |
- |
- |
hypothetical protein CICLE_v10025281mg [Citrus clementina] |
| 15 |
Hb_001433_060 |
0.2178588229 |
- |
- |
hypothetical protein JCGZ_23291 [Jatropha curcas] |
| 16 |
Hb_000680_040 |
0.301480196 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 17 |
Hb_034589_010 |
0.3359666046 |
- |
- |
hypothetical protein MIMGU_mgv1a020753mg, partial [Erythranthe guttata] |
| 18 |
Hb_002485_020 |
0.3414650174 |
- |
- |
metal transport family protein [Populus trichocarpa] |
| 19 |
Hb_003811_010 |
0.3440682597 |
- |
- |
PREDICTED: uncharacterized protein LOC102611294 [Citrus sinensis] |
| 20 |
Hb_040948_020 |
0.3453002292 |
- |
- |
- |