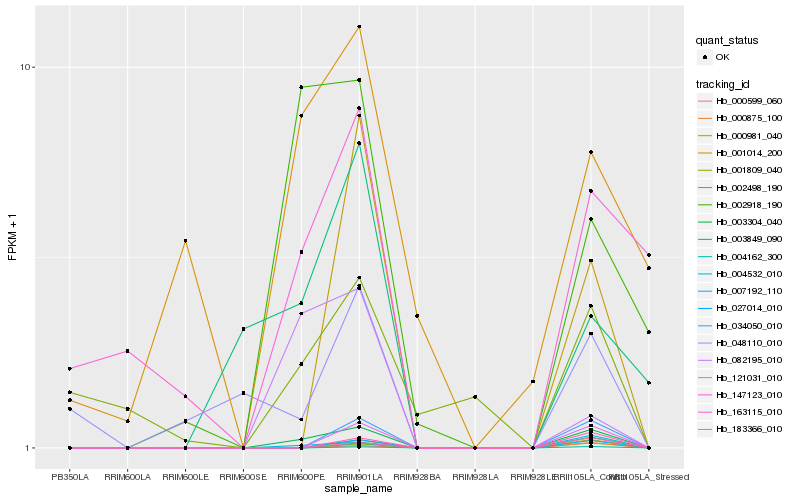
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003304_040 |
0.0 |
- |
- |
hypothetical protein CICLE_v10003343mg, partial [Citrus clementina] |
| 2 |
Hb_007192_110 |
0.0791259964 |
- |
- |
- |
| 3 |
Hb_121031_010 |
0.304751808 |
- |
- |
PREDICTED: uncharacterized protein LOC104104926 [Nicotiana tomentosiformis] |
| 4 |
Hb_034050_010 |
0.3049203668 |
- |
- |
PREDICTED: uncharacterized protein LOC103498182 [Cucumis melo] |
| 5 |
Hb_000875_100 |
0.3102513297 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |
| 6 |
Hb_004162_300 |
0.3135547494 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 7 |
Hb_027014_010 |
0.3154006395 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 8 |
Hb_002918_190 |
0.3203263319 |
- |
- |
Uncharacterized protein TCM_029816 [Theobroma cacao] |
| 9 |
Hb_163115_010 |
0.3271372892 |
- |
- |
- |
| 10 |
Hb_183366_010 |
0.3436286847 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 11 |
Hb_001809_040 |
0.3496902837 |
- |
- |
PREDICTED: uncharacterized protein LOC105638660 [Jatropha curcas] |
| 12 |
Hb_082195_010 |
0.3512797424 |
- |
- |
Uncharacterized protein TCM_043808 [Theobroma cacao] |
| 13 |
Hb_001014_200 |
0.3574301541 |
- |
- |
- |
| 14 |
Hb_000599_060 |
0.3578693869 |
- |
- |
PREDICTED: legumin A-like [Jatropha curcas] |
| 15 |
Hb_003849_090 |
0.3672716446 |
- |
- |
- |
| 16 |
Hb_147123_010 |
0.3699619876 |
- |
- |
Chorismate mutase, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_004532_010 |
0.372157057 |
- |
- |
PREDICTED: uncharacterized protein LOC105800934 [Gossypium raimondii] |
| 18 |
Hb_002498_190 |
0.3745116203 |
- |
- |
- |
| 19 |
Hb_048110_010 |
0.3821455868 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 20 |
Hb_000981_040 |
0.3825389687 |
- |
- |
- |