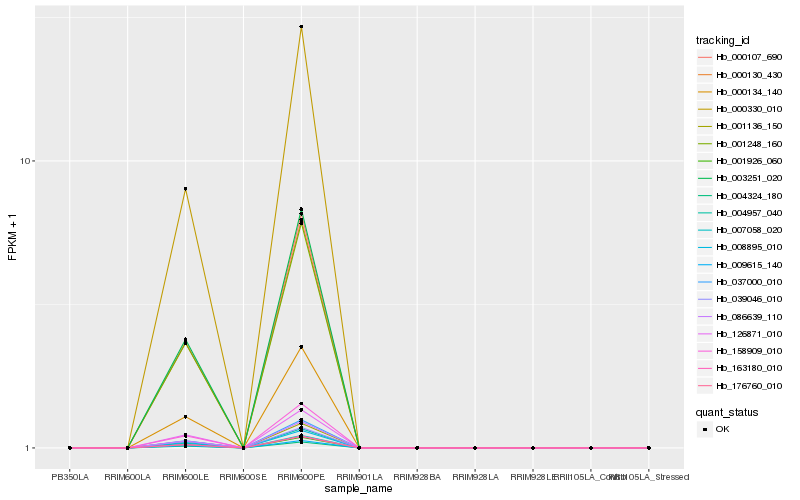
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001248_160 |
0.0 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 2 |
Hb_001136_150 |
0.0036256226 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_163180_010 |
0.0039232486 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 4 |
Hb_000130_430 |
0.0043490627 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 5 |
Hb_001926_060 |
0.0050269572 |
- |
- |
PREDICTED: uncharacterized protein LOC105649615 isoform X2 [Jatropha curcas] |
| 6 |
Hb_007058_020 |
0.0060061113 |
- |
- |
- |
| 7 |
Hb_000330_010 |
0.0093049297 |
- |
- |
hypothetical protein POPTR_0005s25020g [Populus trichocarpa] |
| 8 |
Hb_086639_110 |
0.0094579491 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Theobroma cacao] |
| 9 |
Hb_176760_010 |
0.0132323996 |
- |
- |
PREDICTED: uncharacterized protein LOC105641027 [Jatropha curcas] |
| 10 |
Hb_000107_690 |
0.0137022023 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_003251_020 |
0.0139345902 |
- |
- |
Serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B [Gossypium arboreum] |
| 12 |
Hb_158909_010 |
0.0146555705 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 13 |
Hb_037000_010 |
0.0176463057 |
- |
- |
PREDICTED: putative germin-like protein 2-1 [Jatropha curcas] |
| 14 |
Hb_004957_040 |
0.0194866526 |
- |
- |
PREDICTED: uncharacterized protein LOC104890338 [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_008895_010 |
0.0215890148 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000134_140 |
0.0228142354 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 17 |
Hb_004324_180 |
0.0229310843 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_039046_010 |
0.0281474839 |
- |
- |
PREDICTED: uncharacterized protein LOC105644040 isoform X2 [Jatropha curcas] |
| 19 |
Hb_009615_140 |
0.0297930573 |
- |
- |
PREDICTED: alpha-galactosidase-like isoform X2 [Pyrus x bretschneideri] |
| 20 |
Hb_126871_010 |
0.0301068327 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |