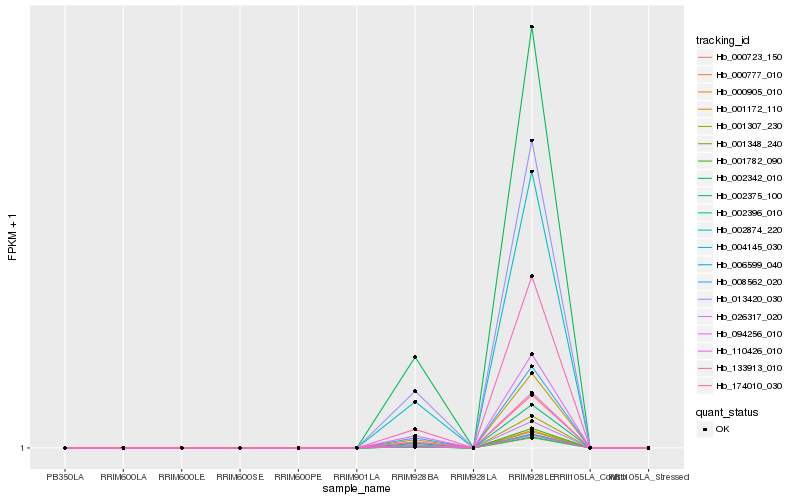
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001172_110 |
0.0 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 2 |
Hb_002375_100 |
0.0008845533 |
- |
- |
PREDICTED: uncharacterized protein LOC104878944 [Vitis vinifera] |
| 3 |
Hb_001307_230 |
0.0018410495 |
- |
- |
PREDICTED: interferon-related developmental regulator 1-like [Jatropha curcas] |
| 4 |
Hb_026317_020 |
0.0030816044 |
- |
- |
hypothetical protein VITISV_035911 [Vitis vinifera] |
| 5 |
Hb_000723_150 |
0.0033907001 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.4-like [Jatropha curcas] |
| 6 |
Hb_110426_010 |
0.0040373297 |
- |
- |
- |
| 7 |
Hb_001782_090 |
0.0046567479 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62910-like [Vitis vinifera] |
| 8 |
Hb_000905_010 |
0.0051225826 |
- |
- |
PREDICTED: cytochrome P450 705A5-like [Jatropha curcas] |
| 9 |
Hb_000777_010 |
0.0053884795 |
- |
- |
PREDICTED: uncharacterized protein LOC105766834 [Gossypium raimondii] |
| 10 |
Hb_006599_040 |
0.0058758296 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 11 |
Hb_004145_030 |
0.0059387947 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 12 |
Hb_174010_030 |
0.0061322944 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 13 |
Hb_002396_010 |
0.0064647336 |
- |
- |
PREDICTED: uncharacterized protein LOC105767002 [Gossypium raimondii] |
| 14 |
Hb_001348_240 |
0.0066237393 |
- |
- |
Putative polyprotein, identical [Solanum demissum] |
| 15 |
Hb_002874_220 |
0.0112278185 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g31250 [Jatropha curcas] |
| 16 |
Hb_008562_020 |
0.011842621 |
- |
- |
- |
| 17 |
Hb_002342_010 |
0.0150275599 |
- |
- |
RALF-LIKE 27 family protein [Populus trichocarpa] |
| 18 |
Hb_094256_010 |
0.0159753042 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_013420_030 |
0.018402826 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_133913_010 |
0.0209781647 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |