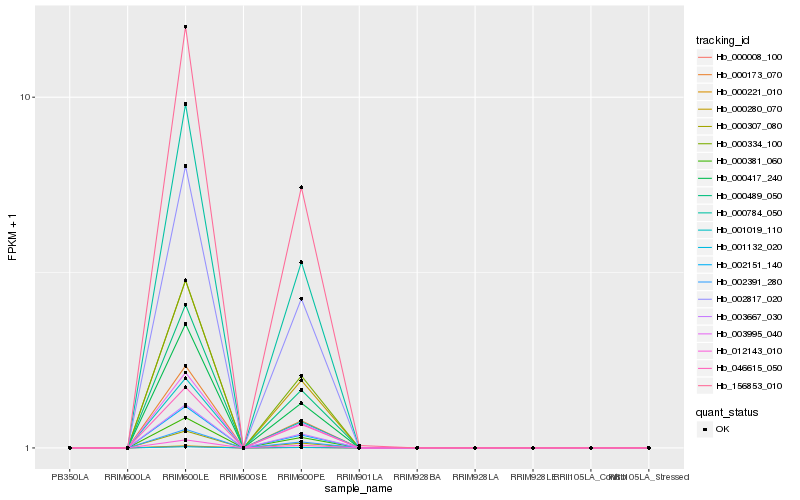
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_156853_010 |
0.0 |
- |
- |
PREDICTED: vinorine synthase-like [Populus euphratica] |
| 2 |
Hb_000334_100 |
0.0204289584 |
- |
- |
hypothetical protein JCGZ_16788 [Jatropha curcas] |
| 3 |
Hb_000008_100 |
0.0204940109 |
- |
- |
Cyclic nucleotide-gated channel 15 [Theobroma cacao] |
| 4 |
Hb_002391_280 |
0.0206360261 |
- |
- |
PREDICTED: ammonium transporter 3 member 1-like [Jatropha curcas] |
| 5 |
Hb_002817_020 |
0.0206972609 |
- |
- |
- |
| 6 |
Hb_000489_050 |
0.0209663705 |
- |
- |
PREDICTED: uncharacterized protein LOC105643984 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003667_030 |
0.0215535557 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL15 [Jatropha curcas] |
| 8 |
Hb_000221_010 |
0.0218262423 |
- |
- |
polyprotein [Oryza australiensis] |
| 9 |
Hb_001019_110 |
0.0230487767 |
- |
- |
PREDICTED: cytochrome P450 734A1 [Jatropha curcas] |
| 10 |
Hb_000381_060 |
0.0232541494 |
- |
- |
- |
| 11 |
Hb_002151_140 |
0.0245890755 |
- |
- |
PREDICTED: lichenase-2-like [Pyrus x bretschneideri] |
| 12 |
Hb_001132_020 |
0.0247396994 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 13 |
Hb_000784_050 |
0.025597194 |
- |
- |
PREDICTED: auxin-induced protein X10A-like [Nelumbo nucifera] |
| 14 |
Hb_000280_070 |
0.0259179198 |
- |
- |
PREDICTED: PCTP-like protein isoform X1 [Jatropha curcas] |
| 15 |
Hb_012143_010 |
0.0265015916 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 [Vitis vinifera] |
| 16 |
Hb_000307_080 |
0.02717331 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000173_070 |
0.0278390052 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor MUTE [Jatropha curcas] |
| 18 |
Hb_003995_040 |
0.0283067551 |
- |
- |
PREDICTED: tropinone reductase homolog At2g29290-like [Jatropha curcas] |
| 19 |
Hb_000417_240 |
0.0284041417 |
- |
- |
PREDICTED: uncharacterized protein LOC105649697 [Jatropha curcas] |
| 20 |
Hb_046615_050 |
0.0287578083 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |