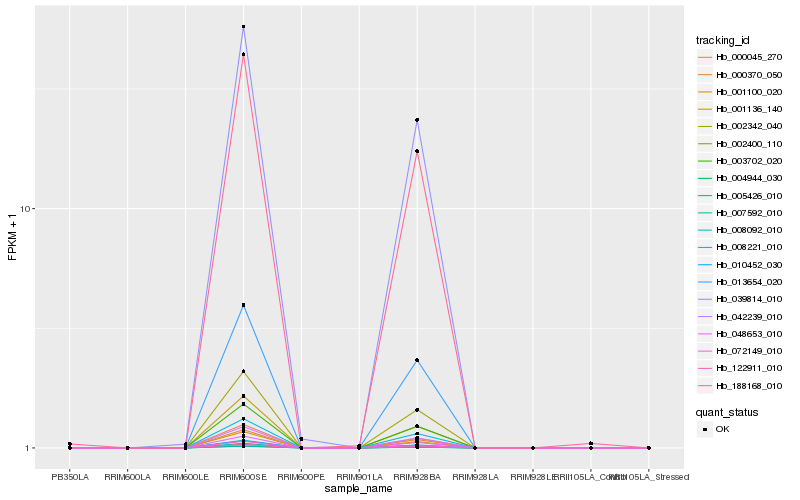
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_122911_010 |
0.0 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000045_270 |
0.0056868766 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002342_040 |
0.0072036307 |
- |
- |
FERONIA, partial [Arabis alpina] |
| 4 |
Hb_010452_030 |
0.0083791505 |
transcription factor |
TF Family: MYB |
Myb domain protein 107 [Theobroma cacao] |
| 5 |
Hb_001100_020 |
0.0096970896 |
- |
- |
- |
| 6 |
Hb_002400_110 |
0.0148779938 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_072149_010 |
0.0151200959 |
- |
- |
hypothetical protein CICLE_v10006890mg, partial [Citrus clementina] |
| 8 |
Hb_004944_030 |
0.0168182033 |
- |
- |
PREDICTED: uncharacterized protein LOC104227769, partial [Nicotiana sylvestris] |
| 9 |
Hb_005426_010 |
0.017589414 |
- |
- |
polyprotein [Oryza australiensis] |
| 10 |
Hb_001136_140 |
0.0176709491 |
- |
- |
hypothetical protein POPTR_0010s13790g [Populus trichocarpa] |
| 11 |
Hb_007592_010 |
0.0177435826 |
- |
- |
PREDICTED: uncharacterized protein LOC105118874 [Populus euphratica] |
| 12 |
Hb_008092_010 |
0.0198898545 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |
| 13 |
Hb_003702_020 |
0.0203336498 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |
| 14 |
Hb_000370_050 |
0.0203691765 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 15 |
Hb_042239_010 |
0.0246464518 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 16 |
Hb_048653_010 |
0.0259087646 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 17 |
Hb_013654_020 |
0.0260509984 |
- |
- |
putative prolin-rich extensin-like receptor protein kinase family protein [Zea mays] |
| 18 |
Hb_039814_010 |
0.0284410897 |
- |
- |
hypothetical protein JCGZ_05023 [Jatropha curcas] |
| 19 |
Hb_008221_010 |
0.0296862716 |
- |
- |
hypothetical protein JCGZ_10961 [Jatropha curcas] |
| 20 |
Hb_188168_010 |
0.030552433 |
- |
- |
Arylacetamide deacetylase, putative [Ricinus communis] |