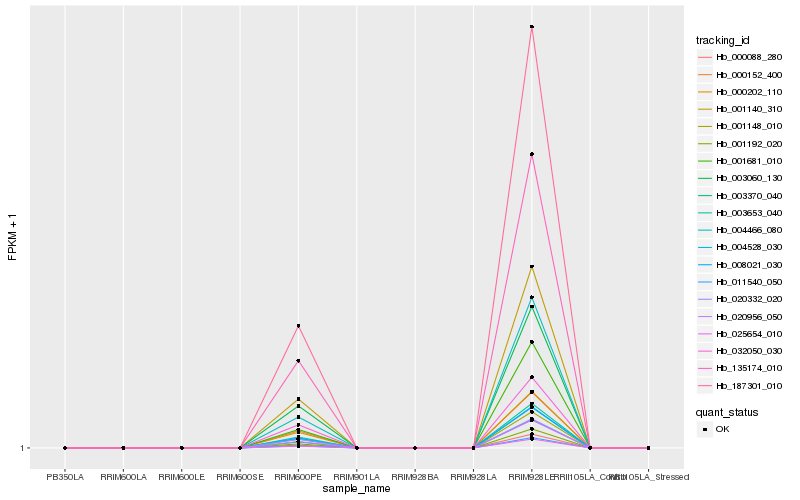
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008021_030 |
0.0 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_001140_310 |
0.0009596454 |
- |
- |
hypothetical protein AMTR_s00025p00025050 [Amborella trichopoda] |
| 3 |
Hb_004528_030 |
0.0009639077 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 4 |
Hb_135174_010 |
0.000988567 |
- |
- |
Glycosyl hydrolase family protein 43 isoform 2 [Theobroma cacao] |
| 5 |
Hb_020956_050 |
0.0030004463 |
- |
- |
PREDICTED: uncharacterized protein LOC102670328 [Glycine max] |
| 6 |
Hb_003653_040 |
0.0055743914 |
- |
- |
hypothetical protein SORBIDRAFT_05g024215 [Sorghum bicolor] |
| 7 |
Hb_001148_010 |
0.0087301878 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 8 |
Hb_001192_020 |
0.0141581734 |
- |
- |
- |
| 9 |
Hb_003370_040 |
0.0158668308 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 10 |
Hb_000088_280 |
0.0202465707 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 11 |
Hb_187301_010 |
0.0237240986 |
- |
- |
PREDICTED: uncharacterized protein LOC105629082 [Jatropha curcas] |
| 12 |
Hb_011540_050 |
0.0240197938 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_003060_130 |
0.0244949302 |
- |
- |
PREDICTED: protein P21-like [Jatropha curcas] |
| 14 |
Hb_000202_110 |
0.0249769067 |
- |
- |
PREDICTED: uncharacterized protein LOC103948916 [Pyrus x bretschneideri] |
| 15 |
Hb_025654_010 |
0.0255388557 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 16 |
Hb_004466_080 |
0.0394611249 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit gamma-1-like [Jatropha curcas] |
| 17 |
Hb_020332_020 |
0.0406130707 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 18 |
Hb_000152_400 |
0.0428775689 |
- |
- |
PREDICTED: uncharacterized protein LOC105648582 [Jatropha curcas] |
| 19 |
Hb_032050_030 |
0.0525620472 |
- |
- |
C, putative [Ricinus communis] |
| 20 |
Hb_001681_010 |
0.0584546672 |
- |
- |
PREDICTED: cytochrome P450 81E8-like isoform X1 [Populus euphratica] |