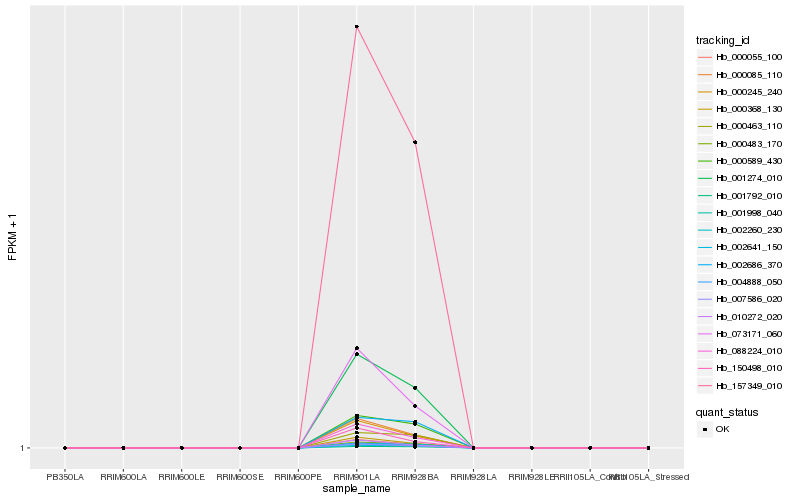
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007586_020 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 2 |
Hb_004888_050 |
0.001611172 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 3 |
Hb_157349_010 |
0.0242160865 |
- |
- |
PREDICTED: uncharacterized protein LOC102588546 [Solanum tuberosum] |
| 4 |
Hb_001274_010 |
0.0331290734 |
- |
- |
- |
| 5 |
Hb_000085_110 |
0.0385197416 |
- |
- |
hypothetical protein JCGZ_00957 [Jatropha curcas] |
| 6 |
Hb_000245_240 |
0.039457004 |
- |
- |
- |
| 7 |
Hb_088224_010 |
0.040700642 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 8 |
Hb_000368_130 |
0.0454153347 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000055_100 |
0.0461856261 |
- |
- |
polyprotein [Oryza australiensis] |
| 10 |
Hb_000483_170 |
0.0574179503 |
transcription factor |
TF Family: NAC |
PREDICTED: protein FEZ [Jatropha curcas] |
| 11 |
Hb_073171_060 |
0.0590105907 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 12 |
Hb_000589_430 |
0.0698131486 |
- |
- |
hypothetical protein JCGZ_01402 [Jatropha curcas] |
| 13 |
Hb_010272_020 |
0.0706425063 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 14 |
Hb_001792_010 |
0.0929210143 |
- |
- |
Uncharacterized protein TCM_043787 [Theobroma cacao] |
| 15 |
Hb_002641_150 |
0.0936816455 |
- |
- |
PREDICTED: stachyose synthase-like [Citrus sinensis] |
| 16 |
Hb_001998_040 |
0.0947531263 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_002260_230 |
0.0952660147 |
- |
- |
hypothetical protein SORBIDRAFT_05g006180 [Sorghum bicolor] |
| 18 |
Hb_150498_010 |
0.0989370106 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 19 |
Hb_000463_110 |
0.1032948154 |
- |
- |
- |
| 20 |
Hb_002686_370 |
0.1035242183 |
- |
- |
PREDICTED: uncharacterized protein LOC102628436 [Citrus sinensis] |