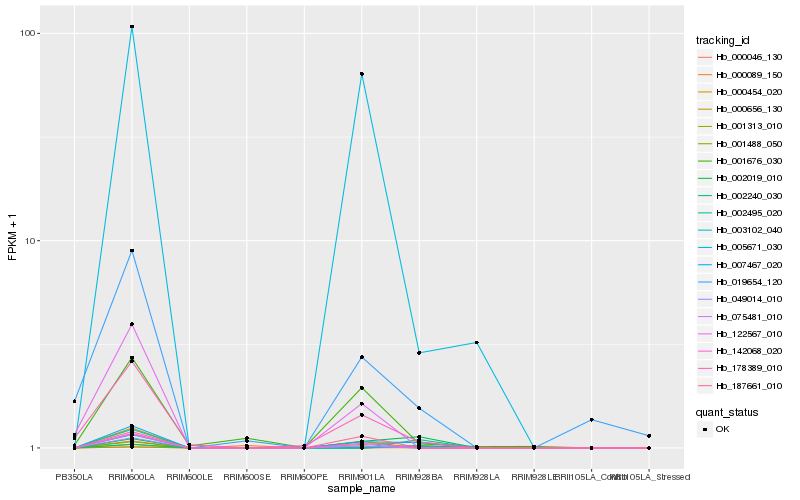
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007467_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_001313_010 |
0.0621329919 |
- |
- |
polyprotein [Ananas comosus] |
| 3 |
Hb_178389_010 |
0.2658650747 |
- |
- |
PREDICTED: U-box domain-containing protein 15-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_019654_120 |
0.274317388 |
- |
- |
PREDICTED: ganglioside-induced differentiation-associated protein 2-like [Jatropha curcas] |
| 5 |
Hb_000656_130 |
0.2819396412 |
- |
- |
hypothetical protein PRUPE_ppa026157mg, partial [Prunus persica] |
| 6 |
Hb_000454_020 |
0.2825683214 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 7 |
Hb_142068_020 |
0.2826941786 |
- |
- |
PREDICTED: thylakoidal processing peptidase 1, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_049014_010 |
0.2849533829 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 9 |
Hb_005671_030 |
0.2899065754 |
- |
- |
- |
| 10 |
Hb_075481_010 |
0.2922583955 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 11 |
Hb_002019_010 |
0.304804836 |
- |
- |
- |
| 12 |
Hb_001676_030 |
0.3140439159 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_122567_010 |
0.3143203088 |
- |
- |
- |
| 14 |
Hb_001488_050 |
0.3148750383 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor 29-like [Jatropha curcas] |
| 15 |
Hb_003102_040 |
0.3152642822 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 23-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_002495_020 |
0.3218327647 |
- |
- |
hypothetical protein EUGRSUZ_A02403, partial [Eucalyptus grandis] |
| 17 |
Hb_187661_010 |
0.3359632432 |
- |
- |
PREDICTED: thylakoidal processing peptidase 1, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_000089_150 |
0.3451468003 |
- |
- |
PREDICTED: uncharacterized protein LOC105635964 [Jatropha curcas] |
| 19 |
Hb_000046_130 |
0.3461843951 |
- |
- |
Peroxidase 27 precursor, putative [Ricinus communis] |
| 20 |
Hb_002240_030 |
0.3480335819 |
- |
- |
- |