| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
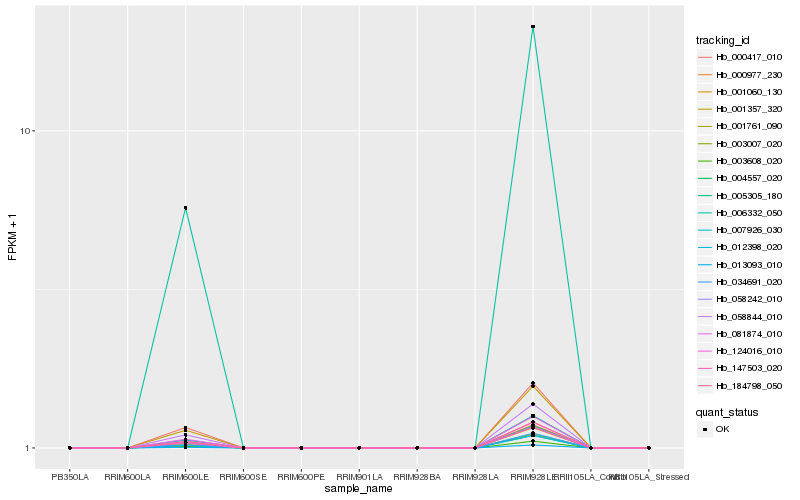
Hb_005305_180 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010-like [Gossypium raimondii] |
| 2 |
Hb_058242_010 |
0.0009793646 |
- |
- |
PREDICTED: uncharacterized protein LOC105793478 [Gossypium raimondii] |
| 3 |
Hb_006332_050 |
0.0010248526 |
- |
- |
hypothetical protein POPTR_0005s07570g [Populus trichocarpa] |
| 4 |
Hb_147503_020 |
0.0015310034 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 5 |
Hb_001761_090 |
0.0024123393 |
- |
- |
Protein STRUBBELIG-RECEPTOR FAMILY 5 [Glycine soja] |
| 6 |
Hb_124016_010 |
0.0051855821 |
- |
- |
hypothetical protein POPTR_0008s21806g [Populus trichocarpa] |
| 7 |
Hb_184798_050 |
0.0056225979 |
- |
- |
- |
| 8 |
Hb_001357_320 |
0.005906636 |
- |
- |
GRP3S, putative [Ricinus communis] |
| 9 |
Hb_007926_030 |
0.0063617991 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP13-like [Populus euphratica] |
| 10 |
Hb_000977_230 |
0.0091300293 |
- |
- |
FT-like protein [Betula luminifera] |
| 11 |
Hb_001060_130 |
0.0124958429 |
- |
- |
- |
| 12 |
Hb_081874_010 |
0.0141117593 |
- |
- |
PREDICTED: uncharacterized protein LOC102581051 [Solanum tuberosum] |
| 13 |
Hb_003608_020 |
0.0180356279 |
- |
- |
hypothetical protein CISIN_1g041945mg [Citrus sinensis] |
| 14 |
Hb_034691_020 |
0.0186315774 |
- |
- |
14 kDa proline-rich protein DC2.15 precursor, putative [Ricinus communis] |
| 15 |
Hb_003007_020 |
0.0192047273 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 16 |
Hb_004557_020 |
0.0207234651 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase V.9-like [Jatropha curcas] |
| 17 |
Hb_012398_020 |
0.0209764766 |
- |
- |
hypothetical protein RCOM_0641040 [Ricinus communis] |
| 18 |
Hb_058844_010 |
0.0216607115 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 19 |
Hb_013093_010 |
0.0218283001 |
- |
- |
retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa Japonica Group] |
| 20 |
Hb_000417_010 |
0.022001316 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase-like [Jatropha curcas] |