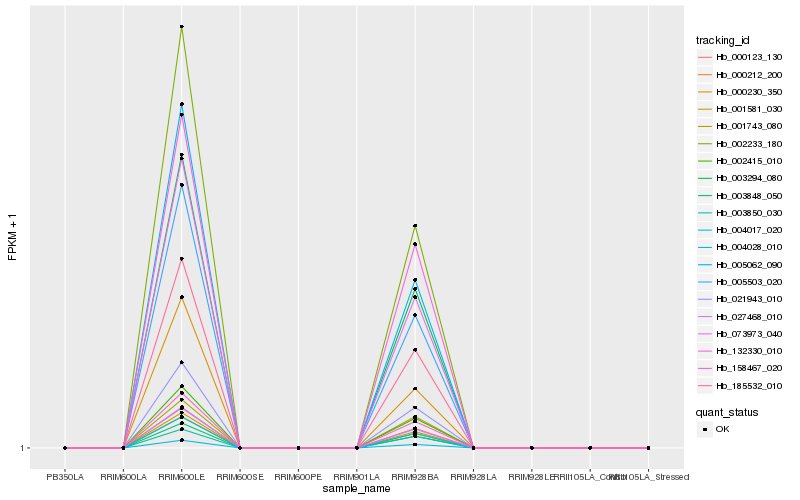
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003294_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_185532_010 |
0.0044225204 |
- |
- |
hypothetical protein POPTR_0006s25570g [Populus trichocarpa] |
| 3 |
Hb_073973_040 |
0.0084172681 |
- |
- |
PREDICTED: uncharacterized protein LOC103941330 [Pyrus x bretschneideri] |
| 4 |
Hb_000123_130 |
0.0097215512 |
- |
- |
hypothetical protein POPTR_0008s05200g, partial [Populus trichocarpa] |
| 5 |
Hb_002233_180 |
0.0118609516 |
- |
- |
- |
| 6 |
Hb_005503_020 |
0.0136393083 |
- |
- |
Hypothetical protein glysoja_025074 [Glycine soja] |
| 7 |
Hb_002415_010 |
0.0139298249 |
- |
- |
PREDICTED: uncharacterized protein LOC105775231 [Gossypium raimondii] |
| 8 |
Hb_158467_020 |
0.0144083587 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 9 |
Hb_003848_050 |
0.0165735679 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |
| 10 |
Hb_004017_020 |
0.0172205939 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 11 |
Hb_021943_010 |
0.0185507327 |
- |
- |
- |
| 12 |
Hb_027468_010 |
0.0210161388 |
- |
- |
kinase, putative [Ricinus communis] |
| 13 |
Hb_001581_030 |
0.0213052695 |
- |
- |
PREDICTED: putative F-box/FBD/LRR-repeat protein At5g56810 [Jatropha curcas] |
| 14 |
Hb_000212_200 |
0.0215171697 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105645015 [Jatropha curcas] |
| 15 |
Hb_004028_010 |
0.0227496042 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 16 |
Hb_005062_090 |
0.0245542393 |
- |
- |
ferredoxin [Vibrio natriegens] |
| 17 |
Hb_132330_010 |
0.0245580714 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003850_030 |
0.0390044222 |
- |
- |
hypothetical protein PRUPE_ppa016115mg [Prunus persica] |
| 19 |
Hb_001743_080 |
0.0481005318 |
- |
- |
- |
| 20 |
Hb_000230_350 |
0.0583047013 |
- |
- |
PREDICTED: uncharacterized protein LOC105630371 [Jatropha curcas] |