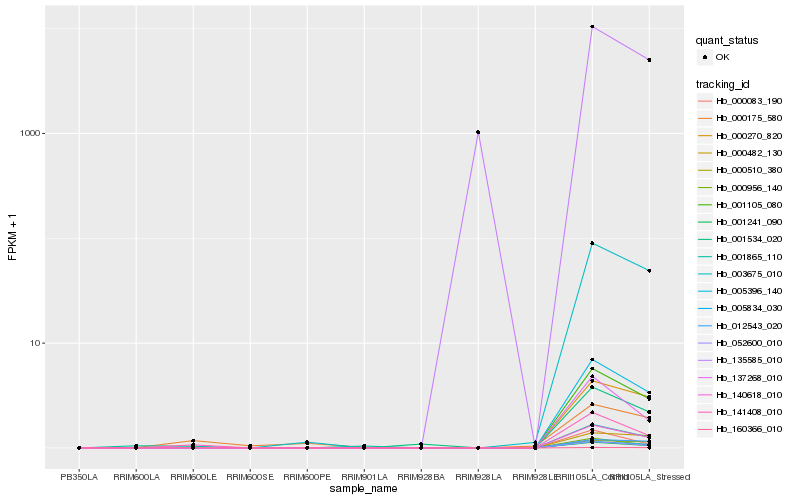
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_141408_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631382 [Jatropha curcas] |
| 2 |
Hb_000956_140 |
0.1512496218 |
- |
- |
- |
| 3 |
Hb_137268_010 |
0.1547623174 |
- |
- |
- |
| 4 |
Hb_000083_190 |
0.1644598198 |
- |
- |
low affinity nitrate transporter, partial [Triticum aestivum] |
| 5 |
Hb_052600_010 |
0.1679373713 |
- |
- |
- |
| 6 |
Hb_001241_090 |
0.1679888776 |
- |
- |
PREDICTED: uncharacterized protein LOC105775709 [Gossypium raimondii] |
| 7 |
Hb_140618_010 |
0.1680027156 |
- |
- |
- |
| 8 |
Hb_005396_140 |
0.1690888662 |
- |
- |
- |
| 9 |
Hb_001105_080 |
0.1717084877 |
- |
- |
- |
| 10 |
Hb_005834_030 |
0.1985462389 |
- |
- |
- |
| 11 |
Hb_001534_020 |
0.203285786 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003675_010 |
0.2048484395 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 13 |
Hb_000270_820 |
0.2199363926 |
- |
- |
- |
| 14 |
Hb_001865_110 |
0.2231568275 |
- |
- |
- |
| 15 |
Hb_000175_580 |
0.2457664661 |
transcription factor, desease resistance |
TF Family: C3H, Gene Name: zf-CCCH |
hypothetical protein POPTR_0013s00890g [Populus trichocarpa] |
| 16 |
Hb_135585_010 |
0.2563850294 |
- |
- |
- |
| 17 |
Hb_160366_010 |
0.2572839857 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_000510_380 |
0.2588872015 |
- |
- |
- |
| 19 |
Hb_000482_130 |
0.2588910505 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 20 |
Hb_012543_020 |
0.2588944807 |
- |
- |
- |